Structural basis of inhibition of CDK-cyclin complexes by INK4 inhibitors
- PMID: 11124804
- PMCID: PMC317144
- DOI: 10.1101/gad.851100
Structural basis of inhibition of CDK-cyclin complexes by INK4 inhibitors
Abstract
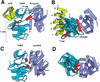
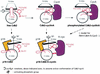
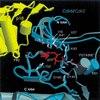
The cyclin-dependent kinases 4 and 6 (Cdk4/6) that drive progression through the G(1) phase of the cell cycle play a central role in the control of cell proliferation, and CDK deregulation is a frequent event in cancer. Cdk4/6 are regulated by the D-type cyclins, which bind to CDKs and activate the kinase, and by the INK4 family of inhibitors. INK4 proteins can bind both monomeric CDK, preventing its association with a cyclin, and also the CDK-cyclin complex, forming an inactive ternary complex. In vivo, binary INK4-Cdk4/6 complexes are more abundant than ternary INK4-Cdk4/6-cyclinD complexes, and it has been suggested that INK4 binding may lead to the eventual dissociation of the cyclin. Here we present the 2.9-A crystal structure of the inactive ternary complex between Cdk6, the INK4 inhibitor p18(INK4c), and a D-type viral cyclin. The structure reveals that p18(INK4c) inhibits the CDK-cyclin complex by distorting the ATP binding site and misaligning catalytic residues. p18(INK4c) also distorts the cyclin-binding site, with the cyclin remaining bound at an interface that is substantially reduced in size. These observations support the model that INK4 binding weakens the cyclin's affinity for the CDK. This structure also provides insights into the specificity of the D-type cyclins for Cdk4/6.
Figures

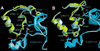

References
-
- Adachi M, Roussel MF, Havenith K, Sherr CJ. Features of macrophage differentiation induced by p19INK4d, a specific inhibitor of cyclin D-dependent kinases. Blood. 1997;90:126–137. - PubMed
-
- Brotherton DH, Dhanaraj V, Wick S, Brizuela L, Domaille PJ, Volyanik E, Xu X, Parisini E, Smith BO, Archer SJ, et al. Crystal structure of the complex of the cyclin D-dependent kinase Cdk6 bound to the cell-cycle inhibitor p19INK4d. Nature. 1998;395:244–250. - PubMed
-
- Brown NR, Noble ME, Endicott JA, Johnson LN. The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat Cell Biol. 1999;1:438–443. - PubMed
-
- Brünger AT, Adams PD, Clore GM, Delano WL, Gros P, Grosse-Kunstieve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
