CluSTr: a database of clusters of SWISS-PROT+TrEMBL proteins
- PMID: 11125042
- PMCID: PMC29804
- DOI: 10.1093/nar/29.1.33
CluSTr: a database of clusters of SWISS-PROT+TrEMBL proteins
Abstract
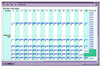
The CluSTr (Clusters of SWISS-PROT and TrEMBL proteins) database offers an automatic classification of SWISS-PROT and TrEMBL proteins into groups of related proteins. The clustering is based on analysis of all pairwise comparisons between protein sequences. Analysis has been carried out for different levels of protein similarity, yielding a hierarchical organisation of clusters. The database provides links to InterPro, which integrates information on protein families, domains and functional sites from PROSITE, PRINTS, Pfam and ProDom. Links to the InterPro graphical interface allow users to see at a glance whether proteins from the cluster share particular functional sites. CluSTr also provides cross-references to HSSP and PDB. The database is available for querying and browsing at http://www.ebi.ac.uk/clustr.
Figures

References
-
- Fleischmann W., Moeller,S., Gateau,A. and Apweiler,R. (1999) A novel method for automatic functional annotation of proteins. Bioinformatics, 15, 228–233. - PubMed
-
- O’Donovan C., Martin,M.J., Glemet,E., Codani,J.J. and Apweiler,R. (1999) Removing redundancy in SWISS-PROT and TrEMBL. Bioinformatics, 15, 258–259. - PubMed
-
- Apweiler R., Biswas,M., Fleischmann,W., Kanapin,A., Karavidopoulou,Y., Kersey,P., Kriventseva,E., Mittard,V., Mulder,N., Phan,I. and Zdobnov,E. (2001) Proteome Analysis Database: online application of InterPro and CluSTr for the functional classification of proteins in whole genomes. Nucleic Acids Res., 29, 44–48. - PMC - PubMed
-
- Smith T.F. and Waterman,M.S. (1981) Identification of common molecular subsequences. J. Mol. Biol., 147, 195–197. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

