Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome
- PMID: 11130730
- PMCID: PMC4377090
- DOI: 10.1038/35048617
Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome
Abstract
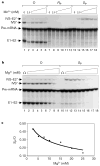
Introns are removed from nuclear messenger RNA precursors through two sequential phospho-transesterification reactions in a dynamic RNA-protein complex called the spliceosome. But whether splicing is catalysed by small nuclear RNAs in the spliceosome is unresolved. As the spliceosome is a metalloenzyme, it is important to determine whether snRNAs coordinate catalytic metals. Here we show that yeast U6 snRNA coordinates a metal ion that is required for the catalytic activity of the spliceosome. With Mg2+, U6 snRNA with a sulphur substitution for the pro-Rp or pro-Sp non-bridging phosphoryl oxygen of nucleotide U80 reconstitutes a fully assembled yet catalytically inactive spliceosome. Adding a thiophilic ion such as Mn2+ allows the first transesterification reaction to occur in the U6/sU80(Sp)- but not the U6/sU80(Rp)-reconstituted spliceosome. Mg2+ competitively inhibits the Mn2+-rescued reaction, indicating that the metal-binding site at U6/U80 exists in the wild-type spliceosome and that the site changes its metal requirement for activity in the Sp spliceosome. Thus, U6 snRNA contributes to pre-messenger RNA splicing through metal-ion coordination, which is consistent with RNA catalysis by the spliceosome.
Figures



Comment in
-
The case for an RNA enzyme.Nature. 2000 Dec 14;408(6814):782-3. doi: 10.1038/35048655. Nature. 2000. PMID: 11130703 No abstract available.
References
-
- Sharp PA. Split genes and RNA splicing. Cell. 1994;77:805–815. - PubMed
-
- Guthrie C. The spliceosome is a dynamic ribonucleoprotein machine. Harvey Lect. 1994;90:59–80. - PubMed
-
- Nilsen TW. In: RNA Structure and Function. Simons R, Grunberg-Manago M, editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 1998. pp. 279–307.
-
- Collins CA, Guthrie C. The question remains: is the spliceosome a ribozyme? Nature Struct Biol. 2000;7:850–854. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

