Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins
- PMID: 11133424
- PMCID: PMC92507
- DOI: 10.1128/AEM.67.1.22-32.2001
Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins
Abstract
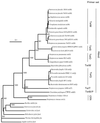
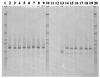
Phylogenetic analysis of tetracycline resistance genes encoding the ribosomal protection proteins (RPPs) revealed the monophyletic origin of these genes. The most deeply branching class, exemplified by tet and otrA, consisted of genes from the antibiotic-producing organisms Streptomyces rimosus and Streptomyces lividans. With a high degree of confidence, the corresponding genes of the other seven classes (Tet M, Tet S, Tet O, Tet W, Tet Q, Tet T, and TetB P) formed phylogenetically distinct separate clusters. Based on this phylogenetic analysis, a set of PCR primers for detection, retrieval, and sequence analysis of the corresponding gene fragments from a variety of bacterial and environmental sources was developed and characterized. A pair of degenerate primers targeted all tetracycline resistance genes encoding RPPs except otrA and tet, and seven other primer pairs were designed to target the specific classes. The primers were used to detect the circulation of these genes in the rumina of cows, in swine feed and feces, and in swine fecal streptococci. Classes Tet O and Tet W were found in the intestinal contents of both animals, while Tet M was confined to pigs and Tet Q was confined to the rumen. The tet(O) and tet(W) genes circulating in the microbiota of the rumen and the gastrointestinal tract of pigs were identical despite the differences in animal hosts and antibiotic use regimens. Swine fecal streptococci uniformly possessed the tet(O) gene, and 22% of them also carried tet(M). This population could be considered one of the main reservoirs of these two resistance genes in the pig gastrointestinal tract. All classes of RPPs except Tet T and TetB P were found in the commercial components of swine feed. This is the first demonstration of the applicability of molecular ecology techniques to estimation of the gene pool and the flux of antibiotic resistance genes in production animals.
Figures




References
-
- Barbosa T M, Scott K P, Flint H J. Evidence for recent intergenic transfer of a new tetracycline resistance gene, tet(W), isolated from Butyrivibrio fibrisolvens, and the occurrence of tet(O), in ruminal bacteria. Environ Microbiol. 1999;1:53–64. - PubMed
-
- Bergeron M G, Quellette M. Diagnosing bacterial infectious diseases in one hour: an essential upcoming revolution. Infection. 1995;23:69–72. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

