Occurrence of Tn4371-related mobile elements and sequences in (chloro)biphenyl-degrading bacteria
- PMID: 11133426
- PMCID: PMC92512
- DOI: 10.1128/AEM.67.1.42-50.2001
Occurrence of Tn4371-related mobile elements and sequences in (chloro)biphenyl-degrading bacteria
Abstract
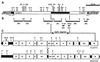
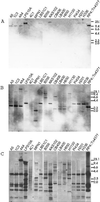
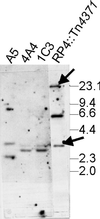
Tn4371, a 55-kb transposable element involved in the degradation and biphenyl or 4-chlorobiphenyl identified in Ralstonia eutropha A5, displays a modular structure including a phage-like integrase gene (int), a Pseudomonas-like (chloro)biphenyl catabolic gene cluster (bph), and RP4- and Ti-plasmid-like transfer genes (trb) (C. Merlin, D. Springael, and A. Toussaint, Plasmid 41:40-54, 1999). Southern blot hybridization was used to examine the presence of different regions of Tn4371 in a collection of (chloro)biphenyl-degrading bacteria originating from different habitats and belonging to different bacterial genera. Tn4371-related sequences were never detected on endogenous plasmids. Although the gene probes containing only bph sequences hybridized to genomic DNA from most strains tested, a limited selection of strains, all beta-proteobacteria, displayed hybridization patterns similar to the Tn4371 bph cluster. Homology between Tn4371 and DNA of two of those strains, originating from the same area as strain A5, extended outside the catabolic genes and covered the putative transfer region of Tn4371. On the other hand, none of the (chloro)biphenyl degraders hybridized with the outer left part of Tn4371 containing the int gene. The bph catabolic determinant of the two strains displaying homology to the Tn4371 transfer genes and a third strain isolated from the A5 area could be mobilized to a R. eutropha recipient, after insertion into an endogenous or introduced IncP1 plasmid. The mobilized DNA of those strains included all Tn4371 homologous sequences previously identified in their genome. Our observations show that the bph genes present on Tn4371 are highly conserved between different (chloro)biphenyl-degrading hosts, isolated globally but belonging mainly to the beta-proteobacteria. On the other hand, Tn4371-related mobile elements carrying bph genes are apparently only found in isolates from the environment that provided the Tn4371-bearing isolate A5.
Figures


References
-
- Ahmad D, Massé R, Sylvestre M. Cloning and expression of genes involved in 4-chlorobiphenyl transformation by Pseudomonas testosteroni: homology to polychlorobiphenyl-degrading genes in other bacteria. Gene. 1990;86:53–61. - PubMed
-
- Asturias J A, Diaz E, Timmis K N. The evolutionary relationship of biphenyl dioxygenase from gram-positive Rhodococcus globerulus P6 to multicomponent dioxygenases from gram-negative bacteria. Gene. 1995;156:11–18. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

