Direct cloning from enrichment cultures, a reliable strategy for isolation of complete operons and genes from microbial consortia
- PMID: 11133432
- PMCID: PMC92523
- DOI: 10.1128/AEM.67.1.89-99.2001
Direct cloning from enrichment cultures, a reliable strategy for isolation of complete operons and genes from microbial consortia
Abstract
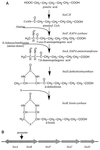
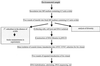
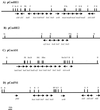
Enrichment cultures of microbial consortia enable the diverse metabolic and catabolic activities of these populations to be studied on a molecular level and to be explored as potential sources for biotechnology processes. We have used a combined approach of enrichment culture and direct cloning to construct cosmid libraries with large (>30-kb) inserts from microbial consortia. Enrichment cultures were inoculated with samples from five environments, and high amounts of avidin were added to the cultures to favor growth of biotin-producing microbes. DNA was extracted from three of these enrichment cultures and used to construct cosmid libraries; each library consisted of between 6,000 and 35,000 clones, with an average insert size of 30 to 40 kb. The inserts contained a diverse population of genomic DNA fragments isolated from the consortia organisms. These three libraries were used to complement the Escherichia coli biotin auxotrophic strain ATCC 33767 Delta(bio-uvrB). Initial screens resulted in the isolation of seven different complementing cosmid clones, carrying biotin biosynthesis operons. Biotin biosynthesis capabilities and growth under defined conditions of four of these clones were studied. Biotin measured in the different culture supernatants ranged from 42 to 3,800 pg/ml/optical density unit. Sequencing the identified biotin synthesis genes revealed high similarities to bio operons from gram-negative bacteria. In addition, random sequencing identified other interesting open reading frames, as well as two operons, the histidine utilization operon (hut), and the cluster of genes involved in biosynthesis of molybdopterin cofactors in bacteria (moaABCDE).
Figures
References
-
- Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. - PubMed
-
- Caldwell D E, Wolfaardts G M, Korber D R, Lawrence J R. Cultivation of microbial consortia and communities. In: Hurst J H, Knudsen G R, McInerney M J, Stetzenbach L D, Walter M V, editors. Manual of environmental microbiology. Washington, D.C.: ASM Press; 1997. pp. 79–90.
-
- Chang Y S, Wu C H, Chang R J, Shiuan D. Determination of biotin concentration by a competitive enzyme-linked immunosorbent assay (ELISA) method. J Biochem, Biophys, Methods. 1994;29:321–329. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

