Phylogenetic specificity and reproducibility and new method for analysis of terminal restriction fragment profiles of 16S rRNA genes from bacterial communities
- PMID: 11133445
- PMCID: PMC92545
- DOI: 10.1128/AEM.67.1.190-197.2001
Phylogenetic specificity and reproducibility and new method for analysis of terminal restriction fragment profiles of 16S rRNA genes from bacterial communities
Abstract
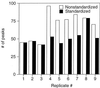
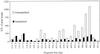
Terminal restriction fragment (TRF) analysis of 16S rRNA genes is an increasingly popular method for rapid comparison of microbial communities, but analysis of the data is still in a developmental stage. We assessed the phylogenetic resolution and reproducibility of TRF profiles in order to evaluate the limitations of the method, and we developed an essential analysis technique to improve the interpretation of TRF data. The theoretical phylogenetic resolution of TRF profiles was determined based on the specificity of TRFs predicted from 3,908 16S rRNA gene sequences. With sequences from the Proteobacteria or gram-positive division, as much as 73% of the TRFs were phylogenetically specific (representing strains from at most two genera). However, the fraction decreased when sequences from the two divisions were combined. The data show that phylogenetic inference will be most effective if TRF profiles represent only a single bacterial division or smaller group. The analytical precision of the TRF method was assessed by comparing nine replicate profiles of a single soil DNA sample. Despite meticulous care in producing the replicates, numerous small, irreproducible peaks were observed. As many as 85% of the 169 distinct TRFs found among the profiles were irreproducible (i.e., not present in all nine replicates). Substantial variation also occurred in the height of synonymous peaks. To make comparisons of microbial communities more reliable, we developed an analytical procedure that reduces variation and extracts a reproducible subset of data from replicate TRF profiles. The procedure can also be used with other DNA fingerprinting techniques for microbial communities or microbial genomes.
Figures


References
-
- Avaniss-Aghajani E, Jones K, Chapman D, Brunk C. A molecular technique for identification of bacteria using small subunit ribosomal RNA sequences. BioTechniques. 1994;17:144–149. - PubMed
-
- Chin K J, Lukow T, Stubner S, Conrad R. Structure and function of the methanogenic archaeal community in stable cellulose-degrading enrichment cultures at two different temperatures (15° and 30°C) FEMS Microbiol Ecol. 1999;30:313–326. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

