Development and optimization of a novel immunomagnetic separation- bacteriophage assay for detection of Salmonella enterica serovar enteritidis in broth
- PMID: 11133448
- PMCID: PMC92550
- DOI: 10.1128/AEM.67.1.217-224.2001
Development and optimization of a novel immunomagnetic separation- bacteriophage assay for detection of Salmonella enterica serovar enteritidis in broth
Abstract
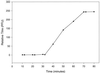
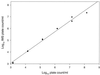
Salmonella is the second-leading cause of food-borne illness in most developed countries, causing diarrhea, cramps, vomiting, and often fever. Many rapid methods are available for detection of Salmonella in foods, but these methods are often insensitive or expensive or require a high degree of technical ability to perform. In this paper we describe development and characterization of a novel assay that utilizes the normal infection cycle of bacteriophage SJ2 for detection of Salmonella enterica serovar Enteritidis in broth. The assay consists of four main stages: (i) capture and concentration of target cells by using immunomagnetic separation (IMS); (ii) infection of the target bacterium with phage; (iii) amplification and recovery of progeny phage; and (iv) assay of progeny phage on the basis of their effect on a healthy population of host cells (signal-amplifying cells). The end point of the assay can be determined by using either fluorescence or optical density measurements. The detection limit of the assay in broth is less than 10(4) CFU/ml, and the assay can be performed in 4 to 5 h. The results of this study demonstrate that the IMS-bacteriophage assay is a rapid, simple, and sensitive technique for detection of Salmonella serovar Enteritidis in broth cultures which can be applied to preenriched food samples.
Figures



References
-
- Bennett A R, Davids F G, Vlahodimou S, Banks J G, Betts R P. The use of bacteriophage-based systems for the separation and concentration of Salmonella. J Appl Microbiol. 1997;83:259–265. - PubMed
-
- Bergey D H, Holt J G, Krieg N R, Sneath P, editors. Bergey's manual of systematic bacteriology. I. Baltimore, Md: Williams and Wilkins; 1984.
-
- Blackburn C D W. Rapid and alternative methods for the detection of salmonellas in foods. J Appl Bacteriol. 1993;75:199–214. - PubMed
-
- Chen J, Griffiths M W. Salmonella detection in eggs using lux+ bacteriophages. J Food Prot. 1996;59:908–914. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

