Chloromethane utilization gene cluster from Hyphomicrobium chloromethanicum strain CM2(T) and development of functional gene probes to detect halomethane-degrading bacteria
- PMID: 11133460
- PMCID: PMC92571
- DOI: 10.1128/AEM.67.1.307-316.2001
Chloromethane utilization gene cluster from Hyphomicrobium chloromethanicum strain CM2(T) and development of functional gene probes to detect halomethane-degrading bacteria
Abstract
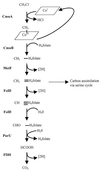
Hyphomicrobium chloromethanicum CM2(T), an aerobic methylotrophic member of the alpha subclass of the class proteobacteria, can grow with chloromethane as the sole carbon and energy source. H. chloromethanicum possesses an inducible enzyme system for utilization of chloromethane, in which two polypeptides (67-kDa CmuA and 35-kDa CmuB) are expressed. Previously, four genes, cmuA, cmuB, cmuC, and purU, were shown to be essential for growth of Methylobacterium chloromethanicum on chloromethane. The cmuA and cmuB genes were used as probes to identify homologs in H. chloromethanicum. A cmu gene cluster (9.5 kb) in H. chloromethanicum contained 10 open reading frames: folD (partial), pduX, orf153, orf207, orf225, cmuB, cmuC, cmuA, fmdB, and paaE (partial). CmuA from H. chloromethanicum (67 kDa) showed high identity to CmuA from M. chloromethanicum and contains an N-terminal methyltransferase domain and a C-terminal corrinoid-binding domain. CmuB from H. chloromethanicum is related to a family of methyl transfer proteins and to the CmuB methyltransferase from M. chloromethanicum. CmuC from H. chloromethanicum shows identity to CmuC from M. chloromethanicum and is a putative methyltransferase. folD codes for a methylene-tetrahydrofolate cyclohydrolase, which may be involved in the C(1) transfer pathway for carbon assimilation and CO(2) production, and paaE codes for a putative redox active protein. Molecular analyses and some preliminary biochemical data indicated that the chloromethane utilization pathway in H. chloromethanicum is similar to the corrinoid-dependent methyl transfer system in M. chloromethanicum. PCR primers were developed for successful amplification of cmuA genes from newly isolated chloromethane utilizers and enrichment cultures.
Figures





Similar articles
-
Chloromethane-dependent expression of the cmu gene cluster of Hyphomicrobium chloromethanicum.Appl Environ Microbiol. 2004 Jul;70(7):4177-86. doi: 10.1128/AEM.70.7.4177-4186.2004. Appl Environ Microbiol. 2004. PMID: 15240299 Free PMC article.
-
Identification of methyl halide-utilizing genes in the methyl bromide-utilizing bacterial strain IMB-1 suggests a high degree of conservation of methyl halide-specific genes in gram-negative bacteria.Appl Environ Microbiol. 2001 Apr;67(4):1959-63. doi: 10.1128/AEM.67.4.1959-1963.2001. Appl Environ Microbiol. 2001. PMID: 11282657 Free PMC article.
-
A corrinoid-dependent catabolic pathway for growth of a Methylobacterium strain with chloromethane.Proc Natl Acad Sci U S A. 1999 Apr 13;96(8):4615-20. doi: 10.1073/pnas.96.8.4615. Proc Natl Acad Sci U S A. 1999. PMID: 10200311 Free PMC article.
-
A review of bacterial methyl halide degradation: biochemistry, genetics and molecular ecology.Environ Microbiol. 2002 Apr;4(4):193-203. doi: 10.1046/j.1462-2920.2002.00290.x. Environ Microbiol. 2002. PMID: 12010126 Review.
-
[The biology of aerobic methylobacteria capable of degrading halomethanes].Mikrobiologiia. 2003 Mar-Apr;72(2):149-60. Mikrobiologiia. 2003. PMID: 12751236 Review. Russian.
Cited by
-
A metagenomic-based survey of microbial (de)halogenation potential in a German forest soil.Sci Rep. 2016 Jun 29;6:28958. doi: 10.1038/srep28958. Sci Rep. 2016. PMID: 27353292 Free PMC article.
-
Fluorescence-based bacterial bioreporter for specific detection of methyl halide emissions in the environment.Appl Environ Microbiol. 2013 Nov;79(21):6561-7. doi: 10.1128/AEM.01738-13. Epub 2013 Aug 16. Appl Environ Microbiol. 2013. PMID: 23956392 Free PMC article.
-
Transfer of a Catabolic Pathway for Chloromethane in Methylobacterium Strains Highlights Different Limitations for Growth with Chloromethane or with Dichloromethane.Front Microbiol. 2016 Jul 19;7:1116. doi: 10.3389/fmicb.2016.01116. eCollection 2016. Front Microbiol. 2016. PMID: 27486448 Free PMC article.
-
Biodegradation of chloromethane by Pseudomonas aeruginosa strain NB1 under nitrate-reducing and aerobic conditions.Appl Environ Microbiol. 2004 Aug;70(8):4629-34. doi: 10.1128/AEM.70.8.4629-4634.2004. Appl Environ Microbiol. 2004. PMID: 15294795 Free PMC article.
-
Phylogenomic analysis of proteins that are distinctive of Archaea and its main subgroups and the origin of methanogenesis.BMC Genomics. 2007 Mar 29;8:86. doi: 10.1186/1471-2164-8-86. BMC Genomics. 2007. PMID: 17394648 Free PMC article.
References
-
- Butler J H. Better budgets for methyl halides? Nature. 2000;403:260–261. - PubMed
-
- Coulter C, Hamilton J T G, McRoberts W C, Kulakov L, Larkin M J, Harper D B. Halomethane:bisulfide/halide ion methyltransferase, an unusual corrinoid enzyme of environmental significance isolated from an aerobic methylotroph using chloromethane as the sole carbon source. Appl Environ Microbiol. 1999;65:4301–4312. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

