Cell cycle programs of gene expression control morphogenetic protein localization
- PMID: 11134078
- PMCID: PMC2150683
- DOI: 10.1083/jcb.151.7.1501
Cell cycle programs of gene expression control morphogenetic protein localization
Abstract
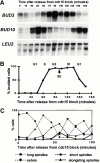
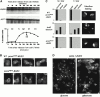
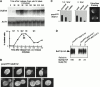
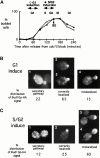
Genomic studies in yeast have revealed that one eighth of genes are cell cycle regulated in their expression. Almost without exception, the significance of cell cycle periodic gene expression has not been tested. Given that many such genes are critical to cellular morphogenesis, we wanted to examine the importance of periodic gene expression to this process. The expression profiles of two genes required for the axial pattern of cell division, BUD3 and BUD10/AXL2/SRO4, are strongly cell cycle regulated. BUD3 is expressed close to the onset of mitosis. BUD10 is expressed in late G1. Through promotor-swap experiments, the expression profile of each gene was altered and the consequences examined. We found that an S/G2 pulse of BUD3 expression controls the timing of Bud3p localization, but that this timing is not critical to Bud3p function. In contrast, a G1 pulse of BUD10 expression plays a direct role in Bud10p localization and function. Bud10p, a membrane protein, relies on the polarized secretory machinery specific to G1 to be delivered to its proper location. Such a secretion-based targeting mechanism for membrane proteins provides cells with flexibility in remodeling their architecture or evolving new forms.
Figures


Similar articles
-
The role of cell cycle-regulated expression in the localization of spatial landmark proteins in yeast.J Cell Biol. 2002 Mar 4;156(5):829-41. doi: 10.1083/jcb.200107041. Epub 2002 Mar 4. J Cell Biol. 2002. PMID: 11877459 Free PMC article.
-
Role of Bud3p in producing the axial budding pattern of yeast.J Cell Biol. 1995 May;129(3):767-78. doi: 10.1083/jcb.129.3.767. J Cell Biol. 1995. PMID: 7730410 Free PMC article.
-
Bud10p directs axial cell polarization in budding yeast and resembles a transmembrane receptor.Curr Biol. 1996 May 1;6(5):570-9. doi: 10.1016/s0960-9822(02)00543-2. Curr Biol. 1996. PMID: 8805277
-
O-Glycosylation of Axl2/Bud10p by Pmt4p is required for its stability, localization, and function in daughter cells.J Cell Biol. 1999 Jun 14;145(6):1177-88. doi: 10.1083/jcb.145.6.1177. J Cell Biol. 1999. PMID: 10366591 Free PMC article.
-
Yeast cell cycle.Curr Opin Cell Biol. 1989 Apr;1(2):250-5. doi: 10.1016/0955-0674(89)90096-3. Curr Opin Cell Biol. 1989. PMID: 2698677 Review. No abstract available.
Cited by
-
Analysis of the landmark protein Bud3 of Ashbya gossypii reveals a novel role in septum construction.EMBO Rep. 2003 Feb;4(2):200-4. doi: 10.1038/sj.embor.embor727. EMBO Rep. 2003. PMID: 12612612 Free PMC article.
-
Central roles of small GTPases in the development of cell polarity in yeast and beyond.Microbiol Mol Biol Rev. 2007 Mar;71(1):48-96. doi: 10.1128/MMBR.00028-06. Microbiol Mol Biol Rev. 2007. PMID: 17347519 Free PMC article. Review.
-
The role of cell cycle-regulated expression in the localization of spatial landmark proteins in yeast.J Cell Biol. 2002 Mar 4;156(5):829-41. doi: 10.1083/jcb.200107041. Epub 2002 Mar 4. J Cell Biol. 2002. PMID: 11877459 Free PMC article.
-
Identification of an amphipathic helix important for the formation of ectopic septin spirals and axial budding in yeast axial landmark protein Bud3p.PLoS One. 2011 Mar 8;6(3):e16744. doi: 10.1371/journal.pone.0016744. PLoS One. 2011. PMID: 21408200 Free PMC article.
-
Septin Assembly and Remodeling at the Cell Division Site During the Cell Cycle.Front Cell Dev Biol. 2021 Nov 25;9:793920. doi: 10.3389/fcell.2021.793920. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34901034 Free PMC article. Review.
References
-
- Chant J., Herskowitz I. Genetic control of bud site selection in yeast by a set of gene products that constitute a morphogenetic pathway. Cell. 1991;65:1203–1212. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

