CD8(+) lymphocytes from simian immunodeficiency virus-infected rhesus macaques recognize 14 different epitopes bound by the major histocompatibility complex class I molecule mamu-A*01: implications for vaccine design and testing
- PMID: 11134287
- PMCID: PMC113970
- DOI: 10.1128/JVI.75.2.738-749.2001
CD8(+) lymphocytes from simian immunodeficiency virus-infected rhesus macaques recognize 14 different epitopes bound by the major histocompatibility complex class I molecule mamu-A*01: implications for vaccine design and testing
Abstract
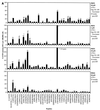
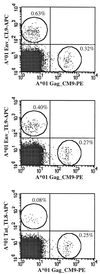
It is becoming increasingly clear that any human immunodeficiency virus (HIV) vaccine should induce a strong CD8(+) response. Additional desirable elements are multispecificity and a focus on conserved epitopes. The use of multiple conserved epitopes arranged in an artificial gene (or EpiGene) is a potential means to achieve these goals. To test this concept in a relevant disease model we sought to identify multiple simian immunodeficiency virus (SIV)-derived CD8(+) epitopes bound by a single nonhuman primate major histocompatibility complex (MHC) class I molecule. We had previously identified the peptide binding motif of Mamu-A*01(2), a common rhesus macaque MHC class I molecule that presents the immunodominant SIV gag-derived cytotoxic T lymphocyte (CTL) epitope Gag_CM9 (CTPYDINQM). Herein, we scanned SIV proteins for the presence of Mamu-A*01 motifs. The binding capacity of 221 motif-positive peptides was determined using purified Mamu-A*01 molecules. Thirty-seven peptides bound with apparent K(d) values of 500 nM or lower, with 21 peptides binding better than the Gag_CM9 peptide. Peripheral blood mononuclear cells from SIV-infected Mamu-A*01(+) macaques recognized 14 of these peptides in ELISPOT, CTL, or tetramer analyses. This study reveals an unprecedented complexity and diversity of anti-SIV CTL responses. Furthermore, it represents an important step toward the design of a multiepitope vaccine for SIV and HIV.
Figures


References
-
- Alexander J, Oseroff C, Sidney J, Wentworth P, Keogh E, Hermanson G, Chisari F V, Kubo R T, Grey H M, Sette A. Derivation of HLA-A11/Kb transgenic mice: functional CTL repertoire and recognition of human A11-restricted CTL epitopes. J Immunol. 1997;159:4753–4761. - PubMed
-
- Allen T M, Sidney J, Delguercio M F, Glickman R L, Lensmeyer G L, Wiebe D A, Demars R, Pauza C D, Johnson R P, Sette A, Watkins D I. Characterization of the peptide binding motif of a rhesus MHC class I molecule (Mamu-A*01) that binds an immunodominant CTL epitope from simian immunodeficiency virus. J Immunol. 1998;160:6062–6071. - PubMed
-
- Allen T M, Vogel T U, Fuller D H, Mothe B R, Steffen S, Boyson J E, Shipley T, Fuller J, Hanke T, Sette A, Altman J D, Moss B, McMichael A J, Watkins D I. Induction of AIDS virus-specific CTL activity in fresh, unstimulated PBL from rhesus macaques vaccinated with a DNA prime/MVA boost regimen. J Immunol. 2000;164:4968–4978. - PubMed
-
- Allen T M, Watkins D I. SIV and SHIV CTL epitopes identified in macaques, p. IV 8–13 ( http://hiv-web.lanl.gov/immunology/) In: Korber B, editor. HIV molecular immunology database 1998. Los Alamos, N. Mex: Los Alamos National Laboratory; 1998.
-
- Altman J D, Moss P A H, Goulder P J R, Barouch D H, McHeyzer-Williams M G, Bell J I, McMichael A J, Davis M M. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

