Characterization of a novel simian immunodeficiency virus from guereza colobus monkeys (Colobus guereza) in Cameroon: a new lineage in the nonhuman primate lentivirus family
- PMID: 11134299
- PMCID: PMC113982
- DOI: 10.1128/JVI.75.2.857-866.2001
Characterization of a novel simian immunodeficiency virus from guereza colobus monkeys (Colobus guereza) in Cameroon: a new lineage in the nonhuman primate lentivirus family
Abstract
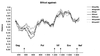
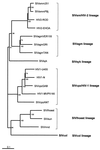
Exploration of the diversity among primate lentiviruses is necessary to elucidate the origins and evolution of immunodeficiency viruses. During a serological survey in Cameroon, we screened 25 wild-born guereza colobus monkeys (Colobus guereza) and identified 7 with HIV/SIV cross-reactive antibodies. In this study, we describe a novel lentivirus, named SIVcol, prevalent in guereza colobus monkeys. Genetic analysis revealed that SIVcol was very distinct from all other known SIV/HIV isolates, with average amino acid identities of 40% for Gag, 50% for Pol, 28% for Env, and around 25% for proteins encoded by five other genes. Phylogenetic analyses confirmed that SIVcol is genetically distinct from other previously characterized primate lentiviruses and clusters independently, forming a novel lineage, the sixth in the current classification. Cercopithecidae monkeys (Old World monkeys) are subdivided into two subfamilies, the Colobinae and the Cercopithecinae, and, so far, all Cercopithecidae monkeys from which lentiviruses have been isolated belong to the Cercopithecinae subfamily. Therefore, SIVcol from guereza colobus monkeys (C. guereza) is the first primate lentivirus identified in the Colobinae subfamily and the divergence of SIVcol may reflect divergence of the host lineage.
Figures




References
-
- Adachi J, Hasegawa M. MOLPHY (a program package for MOLecular PHYlogenetics) version 2.2. Tokyo, Japan: Institute of Statistical Mathematics; 1994.
-
- Beer B E, Bailes E, Goeken R, Dapolito G, Coulibaly C, Norley S G, Kurth R, Gautier J-P, Gautier-Hion A, Vallet D, Sharp P M, Hirsch V M. Simian immunodeficiency virus (SIV) from sun-tailed monkeys (Cercopithecus solatus): evidence for host-dependent evolution of SIV within the C. lhoesti superspecies. J Virol. 1999;73:7734–7744. - PMC - PubMed
-
- Bibollet-Ruche F, Galat-Luong A, Cuny G, Sarni-Manchado P, Galat G, Durand J P, Pourrut X, Veas F. Simian immunodeficiency virus infection in a patas monkey (Erythrocebus patas): evidence for cross-species transmission from African green monkeys (Cercopithecus aethiops sabaeus) in the wild. J Gen Virol. 1996;77:773–781. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

