Analysis of the noncoding regions of measles virus strains in the Edmonston vaccine lineage
- PMID: 11134305
- PMCID: PMC113988
- DOI: 10.1128/JVI.75.2.921-933.2001
Analysis of the noncoding regions of measles virus strains in the Edmonston vaccine lineage
Abstract
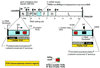
The noncoding sequence of five Edmonston vaccine viruses (AIK-C, Moraten, Rubeovax, Schwarz, and Zagreb) and those of a low-passage Edmonston wild-type (wt) measles virus have been determined and compared. Twenty-one nucleotide positions were identified at which Edmonston wt and one or more vaccine strains differed. The location of some of these nucleotide substitutions suggests that they may influence the efficiency of mRNA synthesis, processing, and translation, as well as genome replication and encapsidation. Five nucleotide substitutions were conserved in all of the vaccine strains. Two of these were in the genomic 3'-terminal transcriptional control region and could affect RNA synthesis or encapsidation. Three were found within the 5'-untranslated region of the F mRNA, potentially altering translation control sequences. The remaining vaccine virus base changes were found in one to four vaccine strains. Their genomic localization suggests that some may modify cis-acting regulatory domains, including the Kozak consensus element of the P and M genes, the F gene-end signal, and the F mRNA 5'-untranslated sequence.
Figures







References
-
- Blumberg B M, Chan J, Udem S A. Function of paramyxovirus 3′ and 5′ end sequences. In: Kingsbury D W, editor. The paramyxoviruses. New York, N.Y: Plenum Press; 1991. pp. 235–247.
-
- Bousse T, Takimoto T, Murti K G, Portner A. Elevated expression of the human parainfluenza virus type 1 F gene downregulates HN expression. Virology. 1997;232:44–52. - PubMed
-
- Cathomen T, Buchholz C J, Spielhofer P, Cattaneo R. Preferential initiation at the second AUG of the measles virus F mRNA: a role for the long untranslated region. Virology. 1995;214:628–632. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

