Coxsackievirus A9 VP1 mutants with enhanced or hindered A particle formation and decreased infectivity
- PMID: 11134308
- PMCID: PMC113991
- DOI: 10.1128/JVI.75.2.952-960.2001
Coxsackievirus A9 VP1 mutants with enhanced or hindered A particle formation and decreased infectivity
Abstract
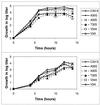
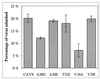
We have studied coxsackievirus A9 (CAV9) mutants that each have a single amino acid substitution in the conserved 29-PALTAVETGHT-39 motif of VP1 and a reduced capacity to produce infectious progeny virus. After uncoating, all steps in the infection cycle occurred according to the same kinetics as and similar efficiency to the wild-type virus. However, the particle/infectious unit ratio in the progeny was significantly increased. The differences were apparently due to altered stability of the capsid: there were mutant viruses with enhanced or hindered uncoating, and both of these characteristics were found to reduce fitness under standard passaging conditions. At 32 degrees C the instable mutants had an advantage, while the wild-type and the most stable mutant grew poorly. When comparing the newly published CAV9 structure and the other enterovirus structures, we found that the PALTAVETGHT motif is always in exactly the same position, in a cavity formed by the 3 other capsid proteins, with the C terminus of VP4 between this motif and the RNA. In the 7 enterovirus structures determined to date, the most conserved residues of the studied motif have identical contacts to neighboring residues of VP2, VP3, and VP4. We conclude that (i) the mutations affect the uncoating step necessary for infection, resulting in an untimely or hindered externalization of the VP1 N terminus together with the VP4, and (ii) the reason for the studied motif being evolutionarily conserved is its role in maintaining an optimal balance between the protective stability and the functional flexibility of the capsid.
Figures


References
-
- Airaksinen A, Roivainen M, Stanway G, Hovi T. Site-saturation mutagenesis of the PALTAVETG motif in coxsackievirus A9 capsid protein VP1 reveals evidence of conservation of a periodic hydrophobicity profile. J Gen Virol. 1999;80:1919–1927. - PubMed
-
- Arnold E, Rossmann M G. The use of molecular-replacement phases for the refinement of the human rhinovirus 14 structure. Acta Crystallogr A. 1988;44:270–282. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

