Association of two novel proteins, TbMP52 and TbMP48, with the Trypanosoma brucei RNA editing complex
- PMID: 11134327
- PMCID: PMC86576
- DOI: 10.1128/MCB.21.2.380-389.2001
Association of two novel proteins, TbMP52 and TbMP48, with the Trypanosoma brucei RNA editing complex
Abstract
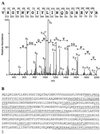
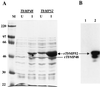
RNA editing in kinetoplastid mitochondria inserts and deletes uridylates at multiple sites in pre-mRNAs as directed by guide RNAs. This occurs by a series of steps that are catalyzed by endoribonuclease, 3'-terminal uridylyl transferase, 3'-exouridylylase, and RNA ligase activities. A multiprotein complex that contains these activities and catalyzes deletion editing in vitro was enriched from Trypanosoma brucei mitochondria by sequential ion-exchange and gel filtration chromatography, followed by glycerol gradient sedimentation. The complex size is approximately 1,600 kDa, and the purified fraction contains 20 major polypeptides. A monoclonal antibody that was generated against the enriched complex reacts with an approximately 49-kDa protein and specifically immunoprecipitates in vitro deletion RNA editing activity. The protein recognized by the antibody was identified by mass spectrometry, and the corresponding gene, designated TbMP52, was cloned. Recombinant TbMP52 reacts with the monoclonal antibody. Another novel protein, TbMP48, which is similar to TbMP52, and its gene were also identified in the enriched complex. These results suggest that TbMP52 and TbMP48 are components of the RNA editing complex.
Figures





References
-
- Blum B, Bakalara N, Simpson L. A model for RNA editing in kinetoplastid mitochondria: “guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell. 1990;60:189–198. - PubMed
-
- Estévez A M, Kierszenbaum F, Wirtz E, Bringaud F, Grunstein J, Simpson L. Knockout of the glutamate dehydrogenase gene in bloodstream Trypanosoma bruceiin culture has no effect on editing of mitochondrial mRNAs. Mol Biochem Parasitol. 1999;100:5–17. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
