Stimulation of CREB binding protein nucleosomal histone acetyltransferase activity by a class of transcriptional activators
- PMID: 11134336
- PMCID: PMC86604
- DOI: 10.1128/MCB.21.2.476-487.2001
Stimulation of CREB binding protein nucleosomal histone acetyltransferase activity by a class of transcriptional activators
Abstract
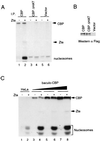
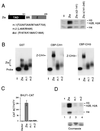
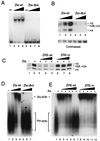
The transcriptional coactivator CREB binding protein (CBP) possesses intrinsic histone acetyltransferase (HAT) activity that is important for gene regulation. CBP binds to and cooperates with numerous nuclear factors to stimulate transcription, but it is unclear if these factors modulate CBP HAT activity. Our previous work showed that CBP interacts with the Epstein-Barr virus-encoded basic region zipper (b-zip) protein, Zta, and augments its transcriptional activity. Here we report that Zta strongly enhances CBP-mediated acetylation of nucleosomal histones. Zta stimulated the HAT activity of CBP that had been partially purified or immunoprecipitated from mammalian cells as well as from affinity-purified, baculovirus expressed CBP. Stimulation of nucleosome acetylation required the CBP HAT domain, the Zta DNA binding and transcription activation domain, and nucleosomal DNA. In addition to Zta, we found that two other b-zip proteins, NF-E2 and C/EBPalpha, strongly stimulated nucleosomal HAT activity. In contrast, several CBP-binding proteins, including phospho-CREB, JUN/FOS, GATA-1, Pit-1, and EKLF, failed to stimulate HAT activity. These results demonstrate that a subset of transcriptional activators enhance the nucleosome-directed HAT activity of CBP and suggest that nuclear factors may regulate transcription by altering substrate recognition and/or the enzymatic activity of chromatin modifying coactivators.
Figures

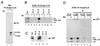


References
-
- Ait-Si-Ali S, Ramirez S, Barre F-X, Dkhissi F, Magnaghi-Jaulin L, Girault J A, Robin P, Knibiehler M, Pritchard L L, Ducommun B, Trouche D, Harel-Bellan A. Histone acetyltransferase activity of CBP is controlled by cycle-dependent kinases and oncoprotein E1A. Nature. 1998;396:184–186. - PubMed
-
- Andersen B, Rosenfeld M G. Pit-1 determines cell types during development of the anterior pituitary gland. A model for transcriptional regulation of cell phenotypes in mammalian organogenesis. J Biol Chem. 1994;269:29335–29338. - PubMed
-
- Andrews N C. The NF-E2 transcription factor. Int J Biochem Cell Biol. 1998;30:429–432. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
