The phosphotyrosyl phosphatase activator, Ncs1p (Rrd1p), functions with Cla4p to regulate the G(2)/M transition in Saccharomyces cerevisiae
- PMID: 11134337
- PMCID: PMC86606
- DOI: 10.1128/MCB.21.2.488-500.2001
The phosphotyrosyl phosphatase activator, Ncs1p (Rrd1p), functions with Cla4p to regulate the G(2)/M transition in Saccharomyces cerevisiae
Abstract
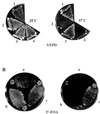
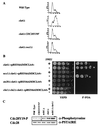
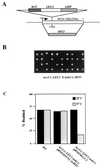
The Saccharomyces cerevisiae p21-activated kinases, Ste20p and Cla4p, have individual functions but appear to share an essential function(s) as well because a strain lacking both kinases is inviable. To learn more about the shared function, we sought new mutations that were lethal in the absence of CLA4. This approach led to the identification of at least 10 complementation groups designated NCS (need CLA4 to survive). As for ste20 cla4-75 mutants, most ncs cla4-75 double mutants were defective for septin localization during budding. One group, NCS1/RRD1 (YIL153w), did not confer this defect, however, and we investigated its function further. ncs1Delta cla4Delta cells arrested with elongated buds and short mitotic spindles. The morphological defects and lethality were suppressed by mutations that abrogate the cell cycle morphogenetic checkpoint, CDC28Y19F or swe1Delta. The connection to the cell cycle may be direct, as we detected a Cla4p-Cdc28p complex. NCS1 encodes a protein with significant similarity to a mammalian phosphotyrosyl phosphatase activator (PTPA) regulatory subunit for type 2A protein phosphatases (PP2As). Genetic and biochemical evidence suggested that the phosphatase Sit4p is a target for Ncs1p. First, CLA4 and SIT4 were synthetically lethal. Second, Ncs1p and its yeast paralog, Noh1p (Rrd2p), bound to the catalytic domain of Sit4p in vitro, and Ncs1p could be immunoprecipitated with Sit4p but not with another PP2A (Pph21p) from yeast cell extracts. Strains lacking both NCS1 and NOH1 were inviable and arrested as unbudded cells, implying that PTPA function is required for proper G(1) progression.
Figures





References
-
- Burns N, Grimwade B, Ross-Macdonald P B, Choi E Y, Finberg K, Roeder G S, Snyder M. Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes Dev. 1994;8:1087–1105. - PubMed
-
- Cayla X, Van Hoof C, Bosch M, Waelkens E, Vandekerckhove J, Peeters B, Merlevede W, Goris J. Molecular cloning, expression, and characterization of PTPA, a protein that activates the tyrosyl phosphatase activity of protein phosphatase 2A. J Biol Chem. 1994;269:15668–15675. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
