Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: Evidence for acetyl transfer from substrate to an NAD breakdown product
- PMID: 11134535
- PMCID: PMC14600
- DOI: 10.1073/pnas.98.2.415
Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: Evidence for acetyl transfer from substrate to an NAD breakdown product
Abstract
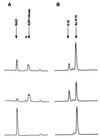
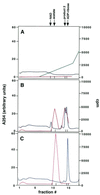
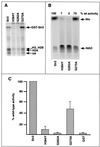
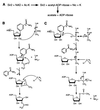
The Saccharomyces cerevisiae silencing protein Sir2 is the founding member of a universally conserved family of proteins that have been shown to possess NAD-dependent histone deacetylation and ADP-ribosylation activities. Here we show that histone deacetylation by Sir2 is coupled to cleavage of the high-energy bond that links the ADP-ribose moiety of NAD to nicotinamide. Analysis of the NAD cleavage products revealed the presence of nicotinamide, ADP-ribose, and a third product that appeared to be related to ADP-ribose. With the use of label transfer experiments, we show that the acetyl group in the histone substrate is transferred to this NAD breakdown product during deacetylation, forming a product that we conclude to be O-acetyl-ADP-ribose. Detection of this species strongly argues for obligate coupling of histone deacetylation to NAD breakdown by Sir2. We propose reaction mechanisms that could account for this coupling via acetyl-ADP-ribose formation. The unprecedented coupling of amide bond cleavage to cleavage of a high-energy bond raises the possibility that NAD breakdown by Sir2 plays an important role in silencing that is independent of its requirement for deacetylation.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

