High-throughput development and characterization of a genomewide collection of gene-based single nucleotide polymorphism markers by chip-based matrix-assisted laser desorption/ionization time-of-flight mass spectrometry
- PMID: 11136232
- PMCID: PMC14630
- DOI: 10.1073/pnas.98.2.581
High-throughput development and characterization of a genomewide collection of gene-based single nucleotide polymorphism markers by chip-based matrix-assisted laser desorption/ionization time-of-flight mass spectrometry
Abstract
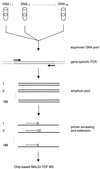
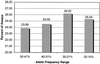
We describe here a system for the rapid identification, assay development, and characterization of gene-based single nucleotide polymorphisms (SNPs). This system couples informatics tools that mine candidate SNPs from public expressed sequence tag resources and automatically designs assay reagents with detection by a chip-based matrix-assisted laser desorption/ionization time-of-flight mass spectrometry platform. As a proof of concept of this system, a genomewide collection of reagents for 9,115 gene-based SNP genetic markers was rapidly developed and validated. These data provide preliminary insights into patterns of polymorphism in a genomewide collection of gene-based polymorphisms.
Figures
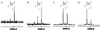
References
-
- Risch N, Merikangas K. Science. 1996;273:1516–1517. - PubMed
-
- Buetow K H, Edmonson M N, Cassidy A B. Nat Genet. 1999;21:323–325. - PubMed
-
- Schuler G D, Boguski M S, Stewart E A, Stein L D, Gyapay G, Rice K, White R E, Rodriguez-Tome P, Aggarwal A, Bajorek E, et al. Science. 1996;274:540–546. - PubMed
-
- Hillier L D, Lennon G, Becker M, Bonaldo M F, Chiapelli B, Chissoe S, Dietrich N, DuBuque T, Favello A, Gish W, et al. Genome Res. 1996;6:807–828. - PubMed
-
- Braun A, Little D P, Koster H. Clin Chem (Washington, DC) 1997;43:1151–1158. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

