Identification of Id4 as a regulator of BRCA1 expression by using a ribozyme-library-based inverse genomics approach
- PMID: 11136250
- PMCID: PMC14556
- DOI: 10.1073/pnas.98.1.130
Identification of Id4 as a regulator of BRCA1 expression by using a ribozyme-library-based inverse genomics approach
Abstract
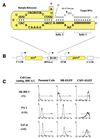
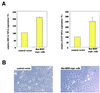
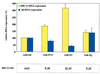
Expression of the breast and ovarian cancer susceptibility gene BRCA1 is down-regulated in sporadic breast and ovarian cancer cases. Therefore, the identification of genes involved in the regulation of BRCA1 expression might lead to new insights into the pathogenesis and treatment of these tumors. In the present study, an "inverse genomics" approach based on a randomized ribozyme gene library was applied to identify cellular genes regulating BRCA1 expression. A ribozyme gene library with randomized target recognition sequences was introduced into human ovarian cancer-derived cells stably expressing a selectable marker [enhanced green fluorescence protein (EGFP)] under the control of the BRCA1 promoter. Cells in which BRCA1 expression was upregulated by particular ribozymes were selected through their concomitant increase in EGFP expression. The cellular target gene of one ribozyme was identified to be the dominant negative transcriptional regulator Id4. Modulation of Id4 expression resulted in inversely regulated expression of BRCA1. In addition, increase in Id4 expression was associated with the ability of cells to exhibit anchorage-independent growth, demonstrating the biological relevance of this gene. Our data suggest that Id4 is a crucial gene regulating BRCA1 expression and might therefore be important for the BRCA1 regulatory pathway involved in the pathogenesis of sporadic breast and ovarian cancer.
Figures


References
-
- Miki Y, Swensen J, Shattuck-Eidens D, Futreal P A, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett L M, Ding W, et al. Science. 1994;266:66–71. - PubMed
-
- Szabo C I, King M C. Hum Mol Genet. 1995;4:1811–1817. - PubMed
-
- Friedman L S, Ostermeyer E A, Szabo C I, Dowd P, Lynch E D, Rowell S E, King M C. Nat Genet. 1994;8:399–404. - PubMed
-
- Smith S A, Easton D F, Evans D G, Ponder B A. Nat Genet. 1992;2:128–131. - PubMed
-
- Futreal P A, Liu Q, Shattuck-Eidens D, Cochran C, Harshman K, Tavtigian S, Bennett L M, Haugen-Strano A, Swensen J, Miki Y, et al. Science. 1994;266:120–122. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

