Variation in the spacer regions separating tRNA genes in Renibacterium salmoninarum distinguishes recent clinical isolates from the same location
- PMID: 11136759
- PMCID: PMC87690
- DOI: 10.1128/JCM.39.1.119-128.2001
Variation in the spacer regions separating tRNA genes in Renibacterium salmoninarum distinguishes recent clinical isolates from the same location
Abstract
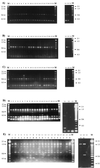
A means for distinguishing between clinical isolates of Renibacterium salmoninarum that is based on the PCR amplification of length polymorphisms in the tRNA intergenic spacer regions (tDNA-ILPs) was investigated. The method used primers specific to nucleotide sequences of R. salmoninarum tRNA genes and tRNA intergenic spacer regions that had been generated by using consensus tRNA gene primers. Twenty-one PCR products were sequenced from five isolates of R. salmoninarum from the United States, England, and Scotland, and four complete tRNA genes and spacer regions were identified. Sixteen specific PCR primers were designed and tested singly and in all possible pairwise combinations for their potential to discriminate between isolates from recent clinical outbreaks of bacterial kidney disease (BKD) in the United Kingdom. Fourteen of the isolates were cultured from kidney samples taken from fish displaying clinical signs of BKD on five farms, and some of the isolates came from the same farm and at the same time. The tDNA-ILP profiles separated 22 clinical isolates into nine groups and highlighted that some farms may have had more than one source of infection. The grouping of isolates improved on the discriminatory power of previously reported typing methods based on randomly amplified polymorphic DNA analysis and restriction fragment length profiles developed using insertion sequence IS994. Our method enabled us to make divisions between closely related clinical isolates of R. salmoninarum that have identical exact tandem repeat (ETR-A) loci, rRNA intergenic spacer sequences, and IS994 profiles.
Figures
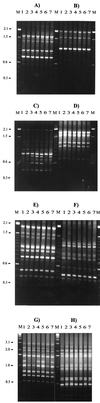

References
-
- Atienzar F, Child P, Evenden A, Jha A, Savva D, Walker C, Depledge M. Application of the arbitrarily primed polymerase chain reaction for the detection of DNA damage. Mar Environ Res. 1998;46:331–335.
-
- Austin B, Embley T M, Goodfellow M. Selective isolation of Renibacterium salmoninarum. FEMS Microbiol Lett. 1983;17:111–114.
-
- Banner C R, Rohovec J S, Fryer J L. A new value for mol percent guanine + cytosine of DNA for the salmonid fish pathogen Renibacterium salmoninarum. FEMS Microbiol Lett. 1991;63:57–59. - PubMed
-
- Bruno D W, Munro A L S. Uniformity in the biochemical properties of Renibacterium salmoninarum isolates obtained from several sources. FEMS Microbiol Lett. 1986;33:247–250.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

