Characterization of Schizosaccharomyces pombe RNA triphosphatase
- PMID: 11139608
- PMCID: PMC29678
- DOI: 10.1093/nar/29.2.387
Characterization of Schizosaccharomyces pombe RNA triphosphatase
Abstract
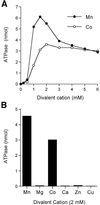
RNA triphosphatase catalyzes the first step in mRNA cap formation which entails the cleavage of the beta-gamma phosphoanhydride bond of triphosphate-terminated RNA to yield a diphosphate end that is then capped with GMP by RNA guanylyltransferase. Here we characterize a 303 amino acid RNA triphosphatase (Pct1p) encoded by the fission yeast SCHIZOSACCHAROMYCES: pombe. Pct1p hydrolyzes the gamma phosphate of triphosphate-terminated poly(A) in the presence of magnesium. Pct1p also hydrolyzes ATP to ADP and P(i) in the presence of manganese or cobalt (K(m) = 19 microM ATP; k(cat) = 67 s(-1)). Hydrolysis of 1 mM ATP is inhibited with increasing potency by inorganic phosphate (I(0.5) = 1 mM), pyrophosphate (I(0.5) = 0.4 mM) and tripolyphosphate (I(0.5) = 30 microM). Velocity sedimentation indicates that Pct1p is a homodimer. Pct1p is biochemically and structurally similar to the catalytic domain of Saccharomyces cerevisiae RNA triphosphatase Cet1p. Mechanistic conservation between Pct1p and Cet1p is underscored by a mutational analysis of the putative metal-binding site of Pct1p. Pct1p is functional in vivo in S.cerevisiae in lieu of Cet1p, provided that it is coexpressed with the S.pombe guanylyltransferase. Pct1p and other yeast RNA triphosphatases are completely unrelated, mechanistically and structurally, to the metazoan RNA triphosphatases, suggesting an abrupt evolutionary divergence of the capping apparatus during the transition from fungal to metazoan species.
Figures

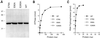





References
-
- Shuman S. (2000) Structure, mechanism and evolution of the mRNA capping apparatus. Prog. Nucleic Acid Res. Mol. Biol., 66, 1–40. - PubMed
-
- Takagi T., Moore,C.R., Diehn,F. and Buratowski,S. (1997) An RNA 5′-triphosphatase related to the protein tyrosine phosphatases. Cell, 89, 867–873. - PubMed
-
- Ho C.K., Sriskanda,V., McCracken,S., Bentley,D., Schwer,B. and Shuman,S. (1998) The guanylyltransferase domain of mammalian mRNA capping enzyme binds to the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem., 273, 9577–9585. - PubMed
-
- Martins A. and Shuman,S. (2000) Mechanism of phosphoanhydride cleavage by baculovirus phosphatase. J. Biol. Chem., 275, 35070–35076. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

