Enzymatic processing of DNA containing tandem dihydrouracil by endonucleases III and VIII
- PMID: 11139610
- PMCID: PMC29670
- DOI: 10.1093/nar/29.2.407
Enzymatic processing of DNA containing tandem dihydrouracil by endonucleases III and VIII
Abstract
Endonuclease III from Escherichia coli, yeast (yNtg1p and yNtg2p) and human and E.coli endonuclease VIII have a wide substrate specificity, and recognize oxidation products of both thymine and cytosine. DNA containing single dihydrouracil (DHU) and tandem DHU lesions were used as substrates for these repair enzymes. It was found that yNtg1p prefers DHU/G and exhibits much weaker enzymatic activity towards DNA containing a DHU/A pair. However, yNtg2p, E. coli and human endonuclease III and E.coli endonuclease VIII activities were much less sensitive to the base opposite the lesion. Although these enzymes efficiently recognize single DHU lesions, they have limited capacity for completely removing this damaged base when DHU is present on duplex DNA as a tandem pair. Both E.coli endonuclease III and yeast yNtg1p are able to remove only one DHU in DNA containing tandem lesions, leaving behind a single DHU at either the 3'- or 5'-terminus of the cleaved fragment. On the other hand, yeast yNtg2p can remove DHU remaining on the 5'-terminus of the 3' cleaved fragment, but is unable to remove DHU remaining on the 3'-terminus of the cleaved 5' fragment. In contrast, both human endonuclease III and E.coli endonuclease VIII can remove DHU remaining on the 3'-terminus of a cleaved 5' fragment, but are unable to remove DHU remaining on the 5'-terminus of a cleaved 3' fragment. Tandem lesions are known to be generated by ionizing radiation and agents that generate reactive oxygen species. The fact that these repair glycosylases have only a limited ability to remove the DHU remaining at the terminus suggests that participation of other repair enzymes is required for the complete removal of tandem lesions before repair synthesis can be efficiently performed by DNA polymerase.
Figures
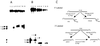

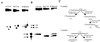

References
-
- Huttermann J., Kuhnlein,W. and Teoule,R. (1978) Effects of Ionizing Radiation on DNA. Physical, Chemical, and Biological Aspects. Springer-Verlag, Berlin. - PubMed
-
- Von Sonntag C. (1987) The Chemical Basis of Radiation Biology. Taylor and Francis, London, UK.
-
- Ward J.F. (1981) Some biochemical consequences of the spatial distribution of ionizing radiation-produced free radicals. Radiat. Res., 86, 185–195. - PubMed
-
- Ward J.F. (1988) DNA damage produced by ionizing radiation in mammalian cells: identities, mechanisms of formation, and repairability. Prog. Nucleic Acids Res. Mol. Biol., 35, 95–125. - PubMed
-
- Ward J.F., Limoli,C.L., Calabrp-Jones,P. and Evans,J.W. (1988) In Nygaard,O.F., Simic,M. and Cerutti,P. (eds), Anticarcinogenesis and Radiation Protection. Plenum, New York, NY.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

