TectoRNA: modular assembly units for the construction of RNA nano-objects
- PMID: 11139616
- PMCID: PMC29663
- DOI: 10.1093/nar/29.2.455
TectoRNA: modular assembly units for the construction of RNA nano-objects
Abstract
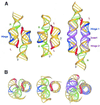
Structural information on complex biological RNA molecules can be exploited to design tectoRNAs or artificial modular RNA units that can self-assemble through tertiary interactions thereby forming nanoscale RNA objects. The selective interactions of hairpin tetraloops with their receptors can be used to mediate tectoRNA assembly. Here we report on the modulation of the specificity and the strength of tectoRNA assembly (in the nanomolar to micromolar range) by variation of the length of the RNA subunits, the nature of their interacting motifs and the degree of flexibility of linker regions incorporated into the molecules. The association is also dependent on the concentration of magnesium. Monitoring of tectoRNA assembly by lead(II) cleavage protection indicates that some degree of structural flexibility is required for optimal binding. With tectoRNAs one can compare the binding affinities of different tertiary motifs and quantify the strength of individual interactions. Furthermore, in analogy to the synthons used in organic chemistry to synthesize more complex organic compounds, tectoRNAs form the basic assembly units for constructing complex RNA structures on the nanometer scale. Thus, tectoRNA provides a means for constructing molecular scaffoldings that organize functional modules in three-dimensional space for a wide range of applications.
Figures


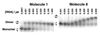

References
-
- Seeman N.C. (1998) DNA nanotechnology: novel DNA constructions. Annu. Rev. Biophys. Biomol. Struct., 27, 225–248. - PubMed
-
- Seeman N.C. (1999) DNA engineering and its application to nanotechnology. Trends Biotechnol., 17, 437–443. - PubMed
-
- Zhang Y. and Seeman,N.C. (1994) The construction of a DNA octahedron. J. Am. Chem. Soc., 116, 1661–1669.
-
- Chen J.H. and Seeman,N.C. (1991) Synthesis from DNA of a molecule with the connectivity of a cube. Nature, 350, 631–633. - PubMed
-
- Winfree E., Liu,F., Wenzler,L.A. and Seeman,N.C. (1998) Design and self-assembly of two-dimensional DNA crystals. Nature, 394, 539–544. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

