DNA-bound transcription factor complexes analysed by mass-spectrometry: binding of novel proteins to the human c-fos SRE and related sequences
- PMID: 11139618
- PMCID: PMC29679
- DOI: 10.1093/nar/29.2.479
DNA-bound transcription factor complexes analysed by mass-spectrometry: binding of novel proteins to the human c-fos SRE and related sequences
Abstract
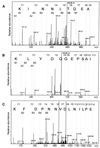
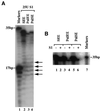
Transcription factors control eukaryotic polymerase II function by influencing the recruitment of multiprotein complexes to promoters and their subsequent integrated function. The complexity of the functional 'transcriptosome' has necessitated biochemical fractionation and subsequent protein sequencing on a grand scale to identify individual components. As a consequence, much is now known of the basal transcription complex. In contrast, less is known about the complexes formed at distal promoter elements. The c-fos SRE, for example, is known to bind Serum Response Factor (SRF) and ternary complex factors such as Elk-1. Their interaction with other factors at the SRE is implied but, to date, none have been identified. Here we describe the use of mass-spectrometric sequencing to identify six proteins, SRF, Elk-1 and four novel proteins, captured on SRE duplexes linked to magnetic beads. This approach is generally applicable to the characterisation of nucleic acid-bound protein complexes and the post-translational modification of their components.
Figures
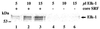




Similar articles
-
Crystal structure of a ternary SAP-1/SRF/c-fos SRE DNA complex.J Mol Biol. 2001 Nov 30;314(3):495-506. doi: 10.1006/jmbi.2001.5138. J Mol Biol. 2001. PMID: 11846562
-
FLI1 and EWS-FLI1 function as ternary complex factors and ELK1 and SAP1a function as ternary and quaternary complex factors on the Egr1 promoter serum response elements.Oncogene. 1997 Jan 16;14(2):213-21. doi: 10.1038/sj.onc.1200839. Oncogene. 1997. PMID: 9010223
-
DNA bending in the ternary nucleoprotein complex at the c-fos promoter.Nucleic Acids Res. 1995 Jul 11;23(13):2442-9. doi: 10.1093/nar/23.13.2442. Nucleic Acids Res. 1995. PMID: 7630721 Free PMC article.
-
Isolation and characterization of SRF accessory proteins.Philos Trans R Soc Lond B Biol Sci. 1993 Jun 29;340(1293):325-32. doi: 10.1098/rstb.1993.0074. Philos Trans R Soc Lond B Biol Sci. 1993. PMID: 8103935 Review.
-
Ets ternary complex transcription factors.Gene. 2004 Jan 7;324:1-14. doi: 10.1016/j.gene.2003.09.028. Gene. 2004. PMID: 14693367 Review.
Cited by
-
Polymorphic potential of SRF binding site of c-Fos gene promoter: in vitro study.RSC Adv. 2024 Dec 3;14(51):38253-38267. doi: 10.1039/d4ra05897f. eCollection 2024 Nov 25. RSC Adv. 2024. PMID: 39628460 Free PMC article.
-
Determining the role of missense mutations in the POU domain of HNF1A that reduce the DNA-binding affinity: A computational approach.PLoS One. 2017 Apr 14;12(4):e0174953. doi: 10.1371/journal.pone.0174953. eCollection 2017. PLoS One. 2017. PMID: 28410371 Free PMC article.
-
Mitogen-induced recruitment of ERK and MSK to SRE promoter complexes by ternary complex factor Elk-1.Nucleic Acids Res. 2008 May;36(8):2594-607. doi: 10.1093/nar/gkn099. Epub 2008 Mar 11. Nucleic Acids Res. 2008. PMID: 18334532 Free PMC article.
-
hCLE/C14orf166 associates with DDX1-HSPC117-FAM98B in a novel transcription-dependent shuttling RNA-transporting complex.PLoS One. 2014 Mar 7;9(3):e90957. doi: 10.1371/journal.pone.0090957. eCollection 2014. PLoS One. 2014. PMID: 24608264 Free PMC article.
-
Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence.Nat Rev Genet. 2011 Dec 6;13(1):59-69. doi: 10.1038/nrg3095. Nat Rev Genet. 2011. PMID: 22143240 Review.
References
-
- Shaw P.E. and Stewart,A.F. (1993) Identification of Protein-DNA Contacts with Dimethyl Sulphate. Humana, Totowa, NJ.
-
- Roberts M.S., Boundy,A., O’Hare,P., Pizzorno,M.C., Ciufo,D.M. and Hayward,G.S. (1988) Direct correlation between a negative autoregulatory response element at the cap site of the herpes simplex virus type 1 IE175 (alpha 4) promoter and a specific binding site for the IE175 (ICP4) protein. J. Virol., 62, 4307–4320. - PMC - PubMed
-
- Franza B.R., Rauscher,F.J.,III, Josephs,S.F. and Curran,T. (1988) The Fos complex and Fos-related antigens recognize sequence elements that contain AP-1 binding sites. Science, 239, 1150–1153. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

