Cis-acting requirements in flanking DNA for the programmed elimination of mse2.9: a common mechanism for deletion of internal eliminated sequences from the developing macronucleus of Tetrahymena thermophila
- PMID: 11139619
- PMCID: PMC29677
- DOI: 10.1093/nar/29.2.488
Cis-acting requirements in flanking DNA for the programmed elimination of mse2.9: a common mechanism for deletion of internal eliminated sequences from the developing macronucleus of Tetrahymena thermophila
Abstract
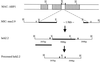
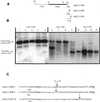
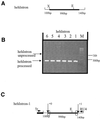
During macronuclear development in the ciliated protozoan Tetrahymena thermophila, extensive DNA deletions occur, eliminating thousands of internal eliminated sequences (IESs). Using an rDNA-based transformation assay we have analyzed the role during DNA deletion of DNA flanking mse2.9, an IES within the second intron of a gene encoding an as yet incompletely characterized protein. We establish that a cis-acting sequence for mse2.9 deletion acts at a distance to specify deletion boundaries. A complex sequence element necessary for efficient and accurate mse2.9 deletion is located in the region 47-81 bp from the right side of mse2.9. The ability of a variety of IES flanking sequences to rescue a processing deficient mse2.9 construct indicates that some cis-acting signal is shared among different IESs. In addition, the short intronic sequence that flanks mse2.9 is able to direct efficient and accurate processing. Despite no obvious sequence similarity between mse2.9 and other IESs, we suggest that a common mechanism is used to delete different families of IESs in Tetrahymena.
Figures





Similar articles
-
Role of micronucleus-limited DNA in programmed deletion of mse2.9 during macronuclear development of Tetrahymena thermophila.Eukaryot Cell. 2004 Apr;3(2):288-301. doi: 10.1128/EC.3.2.288-301.2004. Eukaryot Cell. 2004. PMID: 15075259 Free PMC article.
-
Flanking regulatory sequences of the Tetrahymena R deletion element determine the boundaries of DNA rearrangement.Mol Cell Biol. 1999 Aug;19(8):5631-41. doi: 10.1128/MCB.19.8.5631. Mol Cell Biol. 1999. PMID: 10409752 Free PMC article.
-
Programmed DNA rearrangement from an intron during nuclear development in Tetrahymena thermophila: molecular analysis and identification of potential cis-acting sequences.Nucleic Acids Res. 1996 May 15;24(10):1943-9. doi: 10.1093/nar/24.10.1943. Nucleic Acids Res. 1996. PMID: 8657578 Free PMC article.
-
Developmentally regulated processing and replication of the Tetrahymena rDNA minichromosome.Curr Opin Genet Dev. 1993 Oct;3(5):730-5. doi: 10.1016/s0959-437x(05)80091-7. Curr Opin Genet Dev. 1993. PMID: 8274855 Review.
-
Origin, evolution, and excision of internal elimination segments in germline genes of ciliates.Curr Opin Genet Dev. 1997 Dec;7(6):807-13. doi: 10.1016/s0959-437x(97)80044-5. Curr Opin Genet Dev. 1997. PMID: 9468791 Review.
Cited by
-
A novel family of mobile genetic elements is limited to the germline genome in Tetrahymena thermophila.Nucleic Acids Res. 2002 Jun 1;30(11):2524-37. doi: 10.1093/nar/30.11.2524. Nucleic Acids Res. 2002. PMID: 12034842 Free PMC article.
-
Proteomic Analysis of Histones H2A/H2B and Variant Hv1 in Tetrahymena thermophila Reveals an Ancient Network of Chaperones.Mol Biol Evol. 2019 May 1;36(5):1037-1055. doi: 10.1093/molbev/msz039. Mol Biol Evol. 2019. PMID: 30796450 Free PMC article.
-
Altered tRNA processing is linked to a distinct and unusual La protein in Tetrahymena thermophila.Nat Commun. 2022 Nov 28;13(1):7332. doi: 10.1038/s41467-022-34796-3. Nat Commun. 2022. PMID: 36443289 Free PMC article.
-
Subtraction by addition: domesticated transposases in programmed DNA elimination.Genes Dev. 2009 Nov 1;23(21):2455-60. doi: 10.1101/gad.1864609. Genes Dev. 2009. PMID: 19884252 Free PMC article.
-
Study of an RNA helicase implicates small RNA-noncoding RNA interactions in programmed DNA elimination in Tetrahymena.Genes Dev. 2008 Aug 15;22(16):2228-41. doi: 10.1101/gad.481908. Genes Dev. 2008. PMID: 18708581 Free PMC article.
References
-
- Borst P. and Greaves,D.R. (1987) Programmed gene rearrangements altering gene expression. Science, 235, 658–667. - PubMed
-
- Lewis S.M. (1994) The mechanism of V(D)J joining: lessons from molecular, immunological, and comparative analyses. Adv. Immunol., 56, 27–150. - PubMed
-
- Rudenko G., Cross,M. and Borst,P. (1998) Changing the end: antigenic variation orchestrated at the telomeres of African trypanosomes. Trends Microbiol., 3, 113–117. - PubMed
-
- Klobutcher L.A. and Herrick,G. (1997) Developmental genome reorganization in ciliated protozoa: the transposon link. Prog. Nucleic Acids Res. Mol. Biol., 56, 1–62. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

