A phylogenomic approach to microbial evolution
- PMID: 11139625
- PMCID: PMC29656
- DOI: 10.1093/nar/29.2.545
A phylogenomic approach to microbial evolution
Abstract
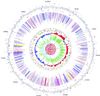
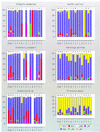
To study the origin and evolution of biochemical pathways in microorganisms, we have developed methods and software for automatic, large-scale reconstructions of phylogenetic relationships. We define the complete set of phylogenetic trees derived from the proteome of an organism as the phylome and introduce the term phylogenetic connection as a concept that describes the relative relationships between taxa in a tree. A query system has been incorporated into the system so as to allow searches for defined categories of trees within the phylome. As a complement, we have developed the pyphy system for visualising the results of complex queries on phylogenetic connections, genomic locations and functional assignments in a graphical format. Our phylogenomics approach, which links phylogenetic information to the flow of biochemical pathways within and among microbial species, has been used to examine more than 8000 phylogenetic trees from seven microbial genomes. The results have revealed a rich web of phylogenetic connections. However, the separation of Bacteria and Archaea into two separate domains remains robust.
Figures



References
-
- Ochman H., Lawrence,J.G. and Groisman,E.A. (2000) Lateral gene transfer and the nature of bacterial innovations. Nature, 405, 299–304. - PubMed
-
- Doolittle W.F. (1999) Phylogenetic classification and the universal tree. Science, 284, 2124–2128. - PubMed
-
- Nelson K.E., Clayton,R.A., Gill,S.R., Gwin,M.L., Dodson,R.J., Haft,D.H., Hickey,E.K., Peterson,J.D., Nelson,W.C., Ketchum,K.A. et al. (1999) Evidence for lateral gene transfer between Archaea and bacteria from the genome sequence of Thermotoga maritima. Nature, 399, 323–329. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

