Efficient recognition of substrates and substrate analogs by the adenine glycosylase MutY requires the C-terminal domain
- PMID: 11139626
- PMCID: PMC29658
- DOI: 10.1093/nar/29.2.553
Efficient recognition of substrates and substrate analogs by the adenine glycosylase MutY requires the C-terminal domain
Abstract
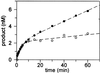
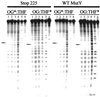
The Escherichia coli DNA repair enzyme MutY plays an important role in the prevention of DNA mutations by removing misincorporated adenine residues from 7, 8-dihydro-8-oxo-2'-deoxyguanosine:2'-deoxyadenosine (OG:A) mispairs. The N-terminal domain of MutY (Stop 225, Met1-Lys225) has a sequence and structure that is characteristic of a superfamily of base excision repair glycosylases; however, MutY and its homologs contain a unique C-terminal domain. Previous studies have shown that the C-terminal domain confers specificity for OG:A substrates over G:A substrates and exhibits homology to the d(OG)TPase MutT, suggesting a role in OG recognition. In order to provide additional information on the importance of the C-terminal domain in damage recognition, we have investigated the kinetic properties of a form lacking this domain (Stop 225) under multiple- and single-turnover conditions. In addition, the interaction of Stop 225 with a series of non-cleavable substrate and product analogs was evaluated using gel retardation assays and footprinting experiments. Under multiple-turnover conditions Stop 225 exhibits biphasic kinetic behavior with both OG:A and G:A substrates, likely due to rate-limiting DNA product release. However, the rate of turnover of Stop 225 was increased 2-fold with OG:A substrates compared to the wild-type enzyme. In contrast, the intrinsic rate for adenine removal by Stop 225 from both G:A and OG:A substrates is significantly reduced (10- to 25-fold) compared to the wild-type. The affinity of Stop 225 for substrate analogs was dramatically reduced, as was the ability to discriminate between substrate analogs paired with OG over G. Interestingly, similar hydroxyl radical and DMS footprinting patterns are observed for Stop 225 and wild-type MutY bound to DNA duplexes containing OG opposite an abasic site mimic or a non-hydrogen bonding A analog, suggesting that similar regions of the DNA are contacted by both enzyme forms. Importantly, Stop 225 has a reduced ability to prevent DNA mutations in vivo. This implies that the reduced adenine glycosylase activity translates to a reduced capacity of Stop 225 to prevent DNA mutations in vivo.
Figures

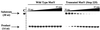

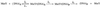
References
-
- Cadet J., Berger,M., Douki,T. and Ravanat,J.L. (1997) Oxidative damage to DNA: formation, measurement and biological significance. Rev. Physiol. Biochem. Pharmacol., 131, 1–87. - PubMed
-
- Henle E.S. and Linn,S. (1997) Formation, prevention and repair of DNA damage by iron/hydrogen peroxide. J. Biol. Chem., 272, 19095–19098. - PubMed
-
- Ames B.N. and Gold,L.S. (1991) Endogenous mutagens and the causes of aging and cancer. Mutat. Res., 250, 121. - PubMed
-
- Gowen L.C., Avrutsky,A.V., Latour,A.M., Koller,B.H. and Leadon,S.A. (1998) BRCA1 required for transcription-coupled repair of oxidative DNA damage. Science, 281, 1009. - PubMed
-
- Lu R., Nash,H.M. and Verdine,G.L. (1997) A mammalian DNA repair enzyme that excises oxidatively damaged guanines maps to a locus frequently lost in lung cancer. Curr. Biol., 7, 397–407. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

