Target DNA chromatinization modulates nicking by L1 endonuclease
- PMID: 11139628
- PMCID: PMC29664
- DOI: 10.1093/nar/29.2.573
Target DNA chromatinization modulates nicking by L1 endonuclease
Abstract
L1 elements are human transposons which replicate via an RNA intermediate. At least 15% of the human genome is composed of L1 sequence. An important initial step in the transposition reaction is nicking of the genomic DNA by L1 endonuclease (L1 EN). In vivo much of the genome exists in the form of chromatin or is undergoing biochemical transactions such as transcription, replication or repair, which may alter the accessibility of the L1 transposition machinery to DNA. To investigate this possibility we have examined the effect of substrate chromatinization on the ability of L1 EN to nick DNA. We find that DNA incorporated into nucleosomes is generally refractory to nicking by L1 EN. Interestingly, nicking of a minority of DNA sequences is enhanced when included in chromatin. Thus, dynamic epigenetic factors such as chromatinization are likely to influence the relatively permanent placement of L1 and other retroelements in the human genome.
Figures




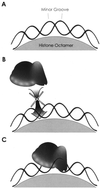
Similar articles
-
Determinants for DNA target structure selectivity of the human LINE-1 retrotransposon endonuclease.Nucleic Acids Res. 2007;35(14):4914-26. doi: 10.1093/nar/gkm516. Epub 2007 Jul 10. Nucleic Acids Res. 2007. PMID: 17626046 Free PMC article.
-
Targeting of human retrotransposon integration is directed by the specificity of the L1 endonuclease for regions of unusual DNA structure.Biochemistry. 1998 Dec 22;37(51):18081-93. doi: 10.1021/bi981858s. Biochemistry. 1998. PMID: 9922177
-
Genome-wide de novo L1 Retrotransposition Connects Endonuclease Activity with Replication.Cell. 2019 May 2;177(4):837-851.e28. doi: 10.1016/j.cell.2019.02.050. Epub 2019 Apr 4. Cell. 2019. PMID: 30955886 Free PMC article.
-
A systematic analysis of LINE-1 endonuclease-dependent retrotranspositional events causing human genetic disease.Hum Genet. 2005 Sep;117(5):411-27. doi: 10.1007/s00439-005-1321-0. Epub 2005 Jun 28. Hum Genet. 2005. PMID: 15983781 Review.
-
Sequence-specific DNA nicking endonucleases.Biomol Concepts. 2015 Aug;6(4):253-67. doi: 10.1515/bmc-2015-0016. Biomol Concepts. 2015. PMID: 26352356 Review.
Cited by
-
Chromatin accessibility: methods, mechanisms, and biological insights.Nucleus. 2022 Dec;13(1):236-276. doi: 10.1080/19491034.2022.2143106. Nucleus. 2022. PMID: 36404679 Free PMC article. Review.
-
A comprehensive analysis of recently integrated human Ta L1 elements.Am J Hum Genet. 2002 Aug;71(2):312-26. doi: 10.1086/341718. Epub 2002 Jun 17. Am J Hum Genet. 2002. PMID: 12070800 Free PMC article.
-
Processed pseudogenes of human endogenous retroviruses generated by LINEs: their integration, stability, and distribution.Genome Res. 2002 Mar;12(3):391-9. doi: 10.1101/gr.216902. Genome Res. 2002. PMID: 11875026 Free PMC article.
-
CCR4, a 3'-5' poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase.EMBO J. 2002 Mar 15;21(6):1414-26. doi: 10.1093/emboj/21.6.1414. EMBO J. 2002. PMID: 11889047 Free PMC article.
-
Viability of X-autosome translocations in mammals: an epigenomic hypothesis from a rodent case-study.Chromosoma. 2004 Aug;113(1):34-41. doi: 10.1007/s00412-004-0292-6. Epub 2004 Jul 9. Chromosoma. 2004. PMID: 15243753
References
-
- Kazazian H.H. and Moran,J.V. (1998) The impact of L1 retrotransposons on the human genome. Nature Genet., 19, 19–24. - PubMed
-
- Moran J.V., Holmes,S.E., Naas,T.P., DeBerardinis,R.J., Boeke,J.D. and Kazazian,H.H.,Jr (1996) High frequency retrotransposition in cultured mammalian cells. Cell, 87, 917–927. - PubMed
-
- Mathias S.L., Scott,A.F., Kazazian,H.H.,Jr, Boeke,J.D. and Gabriel,A. (1991) Reverse transcriptase encoded by a human retrotransposon. Science, 254, 1808–1810. - PubMed
-
- Feng Q., Moran,J., Kazazian,H. and Boeke,J.D. (1996) Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell, 87, 905–916. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

