RNA analysis by ion-pair reversed-phase high performance liquid chromatography
- PMID: 11139637
- PMCID: PMC29688
- DOI: 10.1093/nar/29.2.e7
RNA analysis by ion-pair reversed-phase high performance liquid chromatography
Abstract
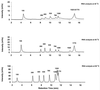
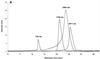
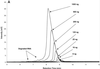
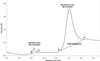
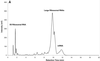
Ion-pair reversed-phase high performance liquid chromatography (IP RP HPLC) is presented as a new, superior method for the analysis of RNA. IP RP HPLC provides a fast and reliable alternative to classical methods of RNA analysis, including separation of different RNA species, quantification and purification. RNA is stable under the analysis conditions used; degradation of RNA during the analyses was not observed. The versatility of IP RP HPLC for RNA analysis is demonstrated. Components of an RNA ladder, ranging in size from 155 to 1770 nt, were resolved. RNA transcripts of up to 5219 nt were analyzed, their integrity determined and they were quantified and purified. Purification of mRNA from total RNA is described, separating mouse rRNA from poly(A)(+) mRNA. IP RP HPLC is also suitable for the separation and purification of DIG-labeled from unlabeled RNA. RNA purified by IP RP HPLC exhibits improved stability.
Figures






Similar articles
-
Two-dimensional reversed-phase x ion-pair reversed-phase HPLC: an alternative approach to high-resolution peptide separation for shotgun proteome analysis.J Proteome Res. 2007 Nov;6(11):4363-73. doi: 10.1021/pr070424t. Epub 2007 Oct 9. J Proteome Res. 2007. PMID: 17924683
-
Purification of poly(A)-messenger ribonucleic acid by reversed-phase high-performance liquid chromatography.J Chromatogr. 1983 Aug 26;266:351-8. doi: 10.1016/s0021-9673(01)90908-2. J Chromatogr. 1983. PMID: 6195171
-
HPLC methods for purity evaluation of man-made single-stranded RNAs.Sci Rep. 2019 Jan 31;9(1):1019. doi: 10.1038/s41598-018-37642-z. Sci Rep. 2019. PMID: 30705318 Free PMC article.
-
Ion-pair reversed-phase chromatography of short double-stranded deoxyribonucleic acid in silicon micro-pillar array columns: retention model and applications.J Chromatogr A. 2013 Jun 14;1294:1-9. doi: 10.1016/j.chroma.2013.04.002. Epub 2013 Apr 8. J Chromatogr A. 2013. PMID: 23647613 Review.
-
A review of chromatographic methods for determination of synthetic food dyes.Talanta. 2010 Jan 15;80(3):1045-51. doi: 10.1016/j.talanta.2009.09.032. Talanta. 2010. PMID: 20006052 Review.
Cited by
-
Purification of linearized template plasmid DNA decreases double-stranded RNA formation during IVT reaction.Front Mol Biosci. 2023 Sep 29;10:1248511. doi: 10.3389/fmolb.2023.1248511. eCollection 2023. Front Mol Biosci. 2023. PMID: 37842641 Free PMC article.
-
Metabolic RNA labeling in non-engineered cells following spontaneous uptake of fluorescent nucleoside phosphate analogues.Nucleic Acids Res. 2024 Sep 23;52(17):10102-10118. doi: 10.1093/nar/gkae722. Nucleic Acids Res. 2024. PMID: 39162218 Free PMC article.
-
Partitioning RNAs by length improves transcriptome reconstruction from short-read RNA-seq data.Nat Biotechnol. 2022 May;40(5):741-750. doi: 10.1038/s41587-021-01136-7. Epub 2022 Jan 10. Nat Biotechnol. 2022. PMID: 35013600 Free PMC article.
-
Rapid purification of RNA secondary structures.Nucleic Acids Res. 2003 Nov 1;31(21):e135. doi: 10.1093/nar/gng136. Nucleic Acids Res. 2003. PMID: 14576335 Free PMC article.
-
Single-nucleotide resolution of RNAs up to 59 nucleotides by high-performance liquid chromatography.Anal Biochem. 2013 Apr 1;435(1):35-43. doi: 10.1016/j.ab.2012.12.011. Epub 2012 Dec 27. Anal Biochem. 2013. PMID: 23274387 Free PMC article.
References
-
- Promega Corp. (1996) Protocols and Applications Guide, 3rd Edn. Promega Corp., Madison, WI, Chap. 6.
-
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, Chap. 7.
-
- Bach H.J., Hartmann,A., Trevors,J.T. and Munch,J.C. (1999) Magnetic capture-hybridization method for purification and probing of mRNA for neutral protease of Bacillus cereus. J. Microbiol. Methods, 37, 187–192. - PubMed
-
- Prescott A.M. and Fricker,C.R. (1999) Use of PNA oligonucleotides for the in situ detection of Escherichia coli in water. Mol. Cell. Probes, 13, 261–268. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

