Extending unbiased stereology of brain ultrastructure to three-dimensional volumes
- PMID: 11141509
- PMCID: PMC134588
- DOI: 10.1136/jamia.2001.0080001
Extending unbiased stereology of brain ultrastructure to three-dimensional volumes
Abstract
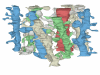
Objective: Analysis of brain ultrastructure is needed to reveal how neurons communicate with one another via synapses and how disease processes alter this communication. In the past, such analyses have usually been based on single or paired sections obtained by electron microscopy. Reconstruction from multiple serial sections provides a much needed, richer representation of the three-dimensional organization of the brain. This paper introduces a new reconstruction system and new methods for analyzing in three dimensions the location and ultrastructure of neuronal components, such as synapses, which are distributed non-randomly throughout the brain.
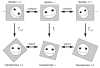
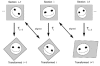
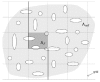
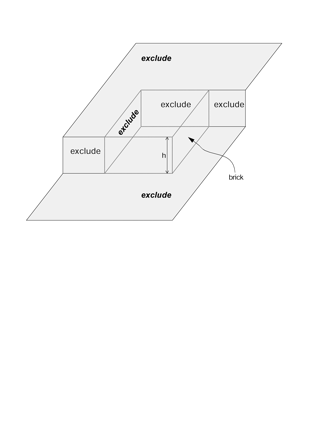
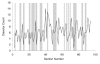
Design and measurements: Volumes are reconstructed by defining transformations that align the entire area of adjacent sections. Whole-field alignment requires rotation, translation, skew, scaling, and second-order nonlinear deformations. Such transformations are implemented by a linear combination of bivariate polynomials. Computer software for generating transformations based on user input is described. Stereological techniques for assessing structural distributions in reconstructed volumes are the unbiased bricking, disector, unbiased ratio, and per-length counting techniques. A new general method, the fractional counter, is also described. This unbiased technique relies on the counting of fractions of objects contained in a test volume. A volume of brain tissue from stratum radiatum of hippocampal area CA1 is reconstructed and analyzed for synaptic density to demonstrate and compare the techniques.
Results and conclusions: Reconstruction makes practicable volume-oriented analysis of ultrastructure using such techniques as the unbiased bricking and fractional counter methods. These analysis methods are less sensitive to the section-to-section variations in counts and section thickness, factors that contribute to the inaccuracy of other stereological methods. In addition, volume reconstruction facilitates visualization and modeling of structures and analysis of three-dimensional relationships such as synaptic connectivity.
Figures








Comment in
-
Human brain program research progress in bioinformatics/ neuroinformatics.J Am Med Inform Assoc. 2001 Jan-Feb;8(1):103-4. doi: 10.1136/jamia.2001.0080103. J Am Med Inform Assoc. 2001. PMID: 11141517 Free PMC article. No abstract available.
References
-
- Jing Z, Sachs F. Alignment of tomographic projections using an incomplete set of fiducial markers. Ultramicroscopy. 1991;35:37–43. - PubMed
-
- Soto GE, Young SJ, Martone ME, et al. Serial section electron tomography: a method for three-dimensional reconstruction of large structures. Neuroimage. 1994;1:230–43. - PubMed
-
- Huijsmans DP, Lamers WH, Los JA, Strackee J. Toward computerized morphometric facilities: a review of 58 software packages for computer-aided three-dimensional reconstruction, quantification, and picture generation from parallel serial sections. Anat Rec. 1986;216:449–70. - PubMed
-
- Stevens JK, Davis TL, Freidman N, Sterling P. A systematic approach to reconstructing microcircuitry by electron microscopy of serial sections. Brain Res Rev. 1980;2:265–93. - PubMed
-
- Stevens JK, Trogadis J. Computer-assisted reconstruction from serial electron micrographs: a tool for the systematic study of neuronal form and function. Adv Cell Neurobiol. 1984;5:341–69.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

