Common data model for neuroscience data and data model exchange
- PMID: 11141510
- PMCID: PMC134589
- DOI: 10.1136/jamia.2001.0080017
Common data model for neuroscience data and data model exchange
Abstract
Objective: Generalizing the data models underlying two prototype neurophysiology databases, the authors describe and propose the Common Data Model (CDM) as a framework for federating a broad spectrum of disparate neuroscience information resources.
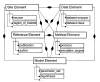
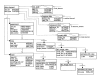
Design: Each component of the CDM derives from one of five superclasses-data, site, method, model, and reference-or from relations defined between them. A hierarchic attribute-value scheme for metadata enables interoperability with variable tree depth to serve specific intra- or broad inter-domain queries. To mediate data exchange between disparate systems, the authors propose a set of XML-derived schema for describing not only data sets but data models. These include biophysical description markup language (BDML), which mediates interoperability between data resources by providing a meta-description for the CDM.
Results: The set of superclasses potentially spans data needs of contemporary neuroscience. Data elements abstracted from neurophysiology time series and histogram data represent data sets that differ in dimension and concordance. Site elements transcend neurons to describe subcellular compartments, circuits, regions, or slices; non-neuroanatomic sites include sequences to patients. Methods and models are highly domain-dependent.
Conclusions: True federation of data resources requires explicit public description, in a metalanguage, of the contents, query methods, data formats, and data models of each data resource. Any data model that can be derived from the defined superclasses is potentially conformant and interoperability can be enabled by recognition of BDML-described compatibilities. Such metadescriptions can buffer technologic changes.
Figures







Comment in
-
Human brain program research progress in bioinformatics/ neuroinformatics.J Am Med Inform Assoc. 2001 Jan-Feb;8(1):103-4. doi: 10.1136/jamia.2001.0080103. J Am Med Inform Assoc. 2001. PMID: 11141517 Free PMC article. No abstract available.
References
-
- Bowden DM, Robertson JE, Martin RF, et al. BrainInfo: a primate brain atlas and neuroanatomic resource based on geographic information system software. Soc Neurosci Abstr. 1998;25:106.
-
- Brinkley JF, Myers LM, Prothero JS, et al. A structural information framework for brain mapping. In: Koslow SH, Huerta MF (eds). Neuroinformatics: An Overview of the Human Brain Project. Mahwah, NJ: Lawrence Erlbaum, 1997:309–34.
-
- Felleman DJ, Van Essen DC. Distributed hierarchical processing in the primate cerebral cortex. Cerebral Cortex. 1991;1:1–47. - PubMed
-
- Mazziotta JC, Toga AW, Evans A, Fox P, Lancaster J. A probabilistic atlas of the human brain: theory and rationale for its development. Neuroimage. 1995;2:89-101. - PubMed
-
- Roland PE, Ziles K. Brain atlases-a new research tool. Trends Neurosci. 1994;17(11):458-467. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

