Integration of multidisciplinary sensory data: a pilot model of the human brain project approach
- PMID: 11141511
- PMCID: PMC134590
- DOI: 10.1136/jamia.2001.0080034
Integration of multidisciplinary sensory data: a pilot model of the human brain project approach
Abstract
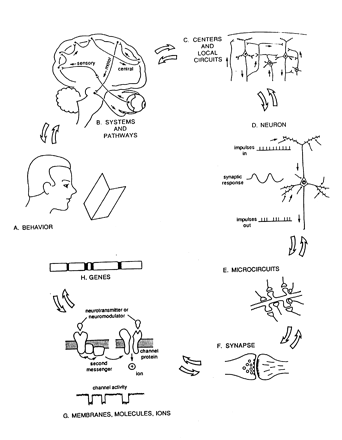
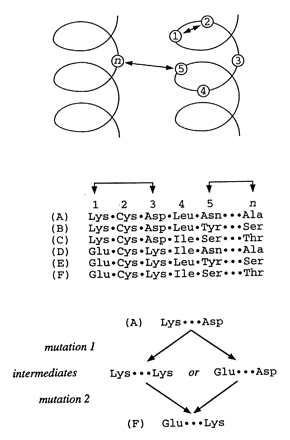
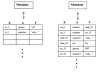
The paper provides an overview of neuroinformatics research at Yale University being performed as part of the national Human Brain Project. This research is exploring the integration of multidisciplinary sensory data, using the olfactory system as a model domain. The neuroinformatics activities fall into three main areas: 1) building databases and related tools that support experimental olfactory research at Yale and can also serve as resources for the field as a whole, 2) using computer models (molecular models and neuronal models) to help understand data being collected experimentally and to help guide further laboratory experiments, 3) performing basic neuroinformatics research to develop new informatics technologies, including a flexible data model (EAV/CR, entity-attribute-value with classes and relationships) designed to facilitate the integration of diverse heterogeneous data within a single unifying framework.
Figures









Comment in
-
Human brain program research progress in bioinformatics/ neuroinformatics.J Am Med Inform Assoc. 2001 Jan-Feb;8(1):103-4. doi: 10.1136/jamia.2001.0080103. J Am Med Inform Assoc. 2001. PMID: 11141517 Free PMC article. No abstract available.
References
-
- Shepherd GM. Neurobiology. New York: Oxford University Press, 1994.
-
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–87. - PubMed
-
- Mori K, Yoshihara Y. Molecular recognition and olfactory processing in the mammalian olfactory system. Prog Neurobiol. 1995;45:585–619. - PubMed
-
- Hildebrand JG, Shepherd GM. Molecular mechanisms of olfactory discrimination: converging evidence for common principles across phyla. Annu Rev Neurosci. 1997;20:595–631. - PubMed
-
- Shepherd GM, Greer CA. Olfactory bulb. In: Shepherd GM (ed). The Synaptic Organization of the Brain. New York: Oxford University Press, 1998.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

