Diagnosis of noncultivatable gastroenteritis viruses, the human caliciviruses
- PMID: 11148001
- PMCID: PMC88960
- DOI: 10.1128/CMR.14.1.15-37.2001
Diagnosis of noncultivatable gastroenteritis viruses, the human caliciviruses
Abstract
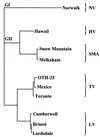
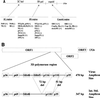
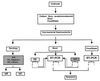
Gastroenteritis is one of the most common illnesses of humans, and many different viruses have been causally associated with this disease. Of those enteric viruses that have been established as etiologic agents of gastroenteritis, only the human caliciviruses cannot be cultivated in vitro. The cloning of Norwalk virus and subsequently of other human caliciviruses has led to the development of several new diagnostic assays. Antigen detection enzyme immunoassays (EIAs) using polyclonal hyperimmune animal sera and antibody detection EIAs using recombinant virus-like particles have supplanted the use of human-derived reagents, but the use of these assays has been restricted to research laboratories. Reverse transcription-PCR assays for the detection of human caliciviruses are more widely available, and these assays have been used to identify virus in clinical specimens as well as in food, water, and other environmental samples. The application of these newer assays has significantly increased the recognition of the importance of human caliciviruses as causes of sporadic and outbreak-associated gastroenteritis.
Figures



References
-
- Adler J L, Zickl R. Winter vomiting disease. J Infect Dis. 1969;119:668–673. - PubMed
-
- Anderson T F, Stanley W M. A study by means of the electron microscope of the reaction between tobacco mosaic virus and its antiserum. J Biol Chem. 1941;139:339–344.
-
- Ando T, Mulders M N, Lewis D C, Estes M K, Monroe S S, Glass R I. Comparison of the polymerase region of small round structured virus strains previously characterized in three serotypes by solid-phase immune electron microscopy. Arch Virol. 1994;135:217–226. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

