Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis
- PMID: 11148285
- PMCID: PMC102225
- DOI: 10.1105/tpc.12.12.2383
Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis
Abstract
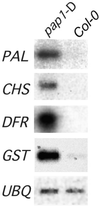
Plants produce a wide array of natural products, many of which are likely to be useful bioactive structures. Unfortunately, these complex natural products usually occur at very low abundance and with restricted tissue distribution, thereby hindering their evaluation. Here, we report a novel approach for enhancing the accumulation of natural products based on activation tagging by Agrobacterium-mediated transformation with a T-DNA that carries cauliflower mosaic virus 35S enhancer sequences at its right border. Among approximately 5000 Arabidopsis activation-tagged lines, we found a plant that exhibited intense purple pigmentation in many vegetative organs throughout development. This upregulation of pigmentation reflected a dominant mutation that resulted in massive activation of phenylpropanoid biosynthetic genes and enhanced accumulation of lignin, hydroxycinnamic acid esters, and flavonoids, including various anthocyanins that were responsible for the purple color. These phenotypes, caused by insertion of the viral enhancer sequences adjacent to an MYB transcription factor gene, indicate that activation tagging can overcome the stringent genetic controls regulating the accumulation of specific natural products during plant development. Our findings suggest a functional genomics approach to the biotechnological evaluation of phytochemical biodiversity through the generation of massively enriched tissue sources for drug screening and for isolating underlying regulatory and biosynthetic genes.
Figures



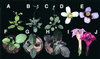
References
-
- Atanassova, R., Foret, N., Martz, F., Chrabbert, B., Tollier, M.-T., Monties, B., Fritig, B., and Legrand, M. (1995). Altered lignin composition in transgenic tobacco expressing O-methyltransferase sequences in sense and antisense orientation. Plant J. 8 465–477.
-
- Bate, N., Orr, J., Ni, W., Meroni, A., Nadler-Hassan, T., Doerner, P.W., Dixon, R.A., Lamb, C.J., and Elkind, Y. (1994). Quantitative relationship between phenylalanine ammonia-lyase levels and phenylpropanoid accumulation in transgenic tobacco identifies a rate-determining step in natural product synthesis. Proc. Natl. Acad. Sci. USA 91 7608–7612. - PMC - PubMed
-
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal Biochem. 72 248–254. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

