Cloning and characterization of IAR1, a gene required for auxin conjugate sensitivity in Arabidopsis
- PMID: 11148286
- PMCID: PMC102226
- DOI: 10.1105/tpc.12.12.2395
Cloning and characterization of IAR1, a gene required for auxin conjugate sensitivity in Arabidopsis
Abstract
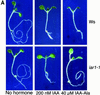
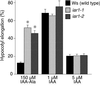
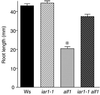
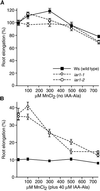
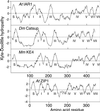
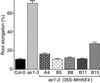
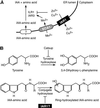
Most indole-3-acetic acid (IAA) in higher plants is conjugated to amino acids, sugars, or peptides, and these conjugates are implicated in regulating the concentration of the free hormone. We identified iar1 as an Arabidopsis mutant that is resistant to the inhibitory effects of several IAA-amino acid conjugates but remains sensitive to free IAA. iar1 partially suppresses phenotypes of a mutant that overproduces IAA, suggesting that IAR1 participates in auxin metabolism or response. We used positional information to clone IAR1, which encodes a novel protein with seven predicted transmembrane domains and several His-rich regions. IAR1 has homologs in other multicellular organisms, including Drosophila, nematodes, and mammals; in addition, the mouse homolog KE4 can functionally substitute for IAR1 in vivo. IAR1 also structurally resembles and has detectable sequence similarity to a family of metal transporters. We discuss several possible roles for IAR1 in auxin homeostasis.
Figures



Similar articles
-
Compensatory mutations in predicted metal transporters modulate auxin conjugate responsiveness in Arabidopsis.G3 (Bethesda). 2013 Jan;3(1):131-41. doi: 10.1534/g3.112.004655. Epub 2013 Jan 1. G3 (Bethesda). 2013. PMID: 23316445 Free PMC article.
-
ILR2, a novel gene regulating IAA conjugate sensitivity and metal transport in Arabidopsis thaliana.Plant J. 2003 Aug;35(4):523-34. doi: 10.1046/j.1365-313x.2003.01826.x. Plant J. 2003. PMID: 12904214
-
An Arabidopsis basic helix-loop-helix leucine zipper protein modulates metal homeostasis and auxin conjugate responsiveness.Genetics. 2006 Dec;174(4):1841-57. doi: 10.1534/genetics.106.061044. Epub 2006 Oct 8. Genetics. 2006. PMID: 17028341 Free PMC article.
-
Auxin: regulation, action, and interaction.Ann Bot. 2005 Apr;95(5):707-35. doi: 10.1093/aob/mci083. Epub 2005 Mar 4. Ann Bot. 2005. PMID: 15749753 Free PMC article. Review.
-
Indole-3-acetic acid protein conjugates: novel players in auxin homeostasis.Plant Biol (Stuttg). 2006 May;8(3):340-5. doi: 10.1055/s-2006-923802. Plant Biol (Stuttg). 2006. PMID: 16807826 Review.
Cited by
-
Comprehensive analysis of the regulatory roles of auxin in early transdifferentiation into xylem cells.Plant Mol Biol. 2009 Jul;70(4):457-69. doi: 10.1007/s11103-009-9485-y. Epub 2009 Mar 27. Plant Mol Biol. 2009. PMID: 19326244
-
Slc39a7/zip7 plays a critical role in development and zinc homeostasis in zebrafish.PLoS One. 2012;7(8):e42939. doi: 10.1371/journal.pone.0042939. Epub 2012 Aug 13. PLoS One. 2012. PMID: 22912764 Free PMC article.
-
Genome-wide association analysis uncovers the genetic architecture of tradeoff between flowering date and yield components in sesame.BMC Plant Biol. 2021 Nov 22;21(1):549. doi: 10.1186/s12870-021-03328-4. BMC Plant Biol. 2021. PMID: 34809568 Free PMC article.
-
Arabidopsis thaliana Yellow Stripe1-Like4 and Yellow Stripe1-Like6 localize to internal cellular membranes and are involved in metal ion homeostasis.Front Plant Sci. 2013 Jul 26;4:283. doi: 10.3389/fpls.2013.00283. eCollection 2013. Front Plant Sci. 2013. PMID: 23898343 Free PMC article.
-
Identification of mouse SLC39A8 as the transporter responsible for cadmium-induced toxicity in the testis.Proc Natl Acad Sci U S A. 2005 Mar 1;102(9):3401-6. doi: 10.1073/pnas.0406085102. Epub 2005 Feb 18. Proc Natl Acad Sci U S A. 2005. PMID: 15722412 Free PMC article.
References
-
- Ando, A., Kikuti, Y.Y., Shigenari, A., Kawata, H., Okamoto, N., Shiina, T., Chen, L., Ikemura, T., Abe, K., Kimura, M., and Inoko, H. (1996). cDNA cloning of the human homologues of the mouse Ke4 and Ke6 genes at the centromeric end of the human MHC region. Genomics 35 600–602. - PubMed
-
- Ausubel, F., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1995). Short Protocols in Molecular Biology. (New York: John Wiley and Sons).
-
- Bandurski, R.S., Cohen, J.D., Slovin, J.P., and Reinecke, D.M. (1995). Auxin biosynthesis and metabolism. In Plant Hormones, P.J. Davies, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 39–65.
-
- Bartel, B. (1997). Auxin biosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48 51–66. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

