PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis
- PMID: 11148287
- PMCID: PMC102227
- DOI: 10.1105/tpc.12.12.2409
PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis
Abstract
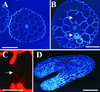
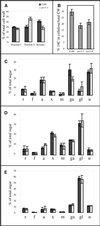
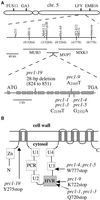
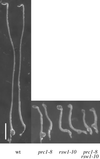
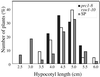
Mutants at the PROCUSTE1 (PRC1) locus show decreased cell elongation, specifically in roots and dark-grown hypocotyls. Cell elongation defects are correlated with a cellulose deficiency and the presence of gapped walls. Map-based cloning of PRC1 reveals that it encodes a member (CesA6) of the cellulose synthase catalytic subunit family, of which at least nine other members exist in Arabidopsis. Mutations in another family member, RSW1 (CesA1), cause similar cell wall defects in all cell types, including those in hypocotyls and roots, suggesting that cellulose synthesis in these organs requires the coordinated expression of at least two distinct cellulose synthase isoforms.
Figures




References
-
- Arioli, T., Peng, L., Betzner, A.S., Burn, J., Wittke, W., Herth, W., Camilleri, C., Hofte, H., Plazinski, J., Birch, R., Cork, A., Glover, J., Redmond, J., and Williamson, R.E. (1998). Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279 717–720. - PubMed
-
- Assaad, F.F., Mayer, U., Wanner, G., and Jurgens, G. (1996). The KEULE gene is involved in cytokinesis in Arabidopsis. Mol. Gen. Genet. 253 267–277. - PubMed
-
- Bichet, A., Desnos, T., Turner, S., Grandjean, O., and Höfte, H. (2001). BOTERO1 is required for normal orientation of cortical microtubules and anisotropic cell expansion in Arabidopsis. Plant J., in press. - PubMed
-
- Bouchez, D., and Camilleri, C. (1998). High molecular weight DNA extraction from Arabidopsis. Methods Mol. Biol. 82 61–70. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

