Mitotic spindle integrity and kinetochore function linked by the Duo1p/Dam1p complex
- PMID: 11149931
- PMCID: PMC2193660
- DOI: 10.1083/jcb.152.1.197
Mitotic spindle integrity and kinetochore function linked by the Duo1p/Dam1p complex
Abstract
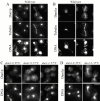
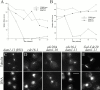
Duo1p and Dam1p were previously identified as spindle proteins in the budding yeast, Saccharomyces cerevisiae. Here, analyses of a diverse collection of duo1 and dam1 alleles were used to develop a deeper understanding of the functions and interactions of Duo1p and Dam1p. Based on the similarity of mutant phenotypes, genetic interactions between duo1 and dam1 alleles, interdependent localization to the mitotic spindle, and Duo1p/Dam1p coimmunoprecipitation from yeast protein extracts, these analyses indicated that Duo1p and Dam1p perform a shared function in vivo as components of a protein complex. Duo1p and Dam1p are not required to assemble bipolar spindles, but they are required to maintain metaphase and anaphase spindle integrity. Immunofluorescence and electron microscopy of duo1 and dam1 mutant spindles revealed a diverse variety of spindle defects. Our results also indicate a second, previously unidentified, role for the Duo1p/Dam1p complex. duo1 and dam1 mutants show high rates of chromosome missegregation, premature anaphase events while arrested in metaphase, and genetic interactions with a subset of kinetochore components consistent with a role in kinetochore function. In addition, Duo1p and Dam1p localize to kinetochores in chromosome spreads, suggesting that this complex may serve as a link between the kinetochore and the mitotic spindle.
Figures




References
-
- Ayscough K.R., Drubin D.G. Immunofluorescence microscopy of yeast cells. In: Celis J., editor. Cell BiologyA Laboratory Handbook. Vol. 2. Academic Press; New York: 1998. pp. 477–485.
-
- Botstein D., Amberg D., Mulholland J., Huffaker T., Adams A., Drubin D., Stearns T. The yeast cytoskeleton. In: Pringle J.R., Broach J.R., Jones E.W., editors. The Molecular and Cellular Biology of the Yeast Saccharomyces. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. pp. 1–90.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

