Structure and dynamics of the GABA binding pocket: A narrowing cleft that constricts during activation
- PMID: 11150321
- PMCID: PMC6762441
- DOI: 10.1523/JNEUROSCI.21-01-00067.2001
Structure and dynamics of the GABA binding pocket: A narrowing cleft that constricts during activation
Abstract
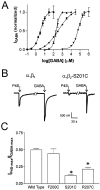
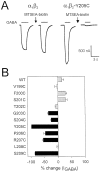
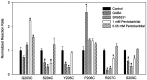
Photo-affinity labeling and mutagenesis studies have identified several amino acids that may contribute to the ligand binding domains of ligand-gated ion channels. These types of studies, however, only generate a one-dimensional, static description of binding site structure. In this study, we used the substituted cysteine accessibility method not only to identify binding pocket residues but also to elicit information about binding site dynamics and structure. Residues surrounding the putative loop C ligand binding domain of the GABA(A) receptor (beta(2)V199 to beta(2)S209) were individually mutated to cysteine, and the mutant subunits were coexpressed with wild-type alpha(1) subunits in Xenopus oocytes. N-biotinylaminoethyl methanethiosulfonate (MTSEA-biotin) reacts with cysteines introduced at positions G203, S204, Y205, P206, R207, and S209. This accessibility pattern is not consistent with either an alpha-helix or beta-strand. Instead, G203-S209 seems to form a water-accessible extended coil, whereas V199-T202 appears to buried in the protein or membrane. Coapplication of either GABA or the competitive antagonist SR-95531 significantly slows MTSEA-biotin modification of cysteines introduced at positions S204, Y205, R207, and S209, demonstrating that these residues line and face into the GABA binding pocket. MTSEA-biotin reaction rates reveal a steep accessibility gradient from G203-S209 and suggests that the binding pocket is a deep narrowing cleft. Pentobarbital activation of the receptor significantly slows MTSEA-biotin modification of cysteines at S204, R207, and S209, suggesting that the binding site may constrict during gating.
Figures


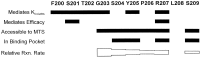
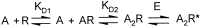
Similar articles
-
A (beta)-strand in the (gamma)2 subunit lines the benzodiazepine binding site of the GABA A receptor: structural rearrangements detected during channel gating.J Neurosci. 2001 Jul 15;21(14):4977-86. doi: 10.1523/JNEUROSCI.21-14-04977.2001. J Neurosci. 2001. PMID: 11438573 Free PMC article.
-
Individually monitoring ligand-induced changes in the structure of the GABAA receptor at benzodiazepine binding site and non-binding-site interfaces.Mol Pharmacol. 2008 Jul;74(1):203-12. doi: 10.1124/mol.108.044891. Epub 2008 Apr 18. Mol Pharmacol. 2008. PMID: 18424553 Free PMC article.
-
Mapping the agonist binding site of the GABAA receptor: evidence for a beta-strand.J Neurosci. 1999 Jun 15;19(12):4847-54. doi: 10.1523/JNEUROSCI.19-12-04847.1999. J Neurosci. 1999. PMID: 10366619 Free PMC article.
-
Combining Mutations and Electrophysiology to Map Anesthetic Sites on Ligand-Gated Ion Channels.Methods Enzymol. 2018;602:369-389. doi: 10.1016/bs.mie.2018.01.014. Epub 2018 Feb 28. Methods Enzymol. 2018. PMID: 29588039 Free PMC article. Review.
-
A closer look at the high affinity benzodiazepine binding site on GABAA receptors.Curr Top Med Chem. 2011;11(2):241-6. doi: 10.2174/156802611794863562. Curr Top Med Chem. 2011. PMID: 21189125 Review.
Cited by
-
Molecular modeling of the GABA(C) receptor ligand-binding domain.J Mol Model. 2006 Feb;12(3):317-24. doi: 10.1007/s00894-005-0034-6. Epub 2005 Oct 26. J Mol Model. 2006. PMID: 16249935
-
Tyrosine residues that control binding and gating in the 5-hydroxytryptamine3 receptor revealed by unnatural amino acid mutagenesis.J Neurosci. 2004 Oct 13;24(41):9097-104. doi: 10.1523/JNEUROSCI.2429-04.2004. J Neurosci. 2004. PMID: 15483128 Free PMC article.
-
Structural insights into the interaction between gabazine (SR-95531) and Laodelphax striatellus GABA receptors.J Pestic Sci. 2022 May 20;47(2):78-85. doi: 10.1584/jpestics.D22-007. J Pestic Sci. 2022. PMID: 35800394 Free PMC article.
-
Principles of agonist recognition in Cys-loop receptors.Front Physiol. 2014 Apr 24;5:160. doi: 10.3389/fphys.2014.00160. eCollection 2014. Front Physiol. 2014. PMID: 24795655 Free PMC article. Review.
-
Rosmarinic Acid, a Bioactive Phenolic Compound, Inhibits Glutamate Release from Rat Cerebrocortical Synaptosomes through GABAA Receptor Activation.Biomolecules. 2021 Jul 15;11(7):1029. doi: 10.3390/biom11071029. Biomolecules. 2021. PMID: 34356653 Free PMC article.
References
-
- Amin J, Weiss DS. GABAA receptor needs two homologous domains of the beta-subunit for activation by GABA but not by pentobarbital. Nature. 1993;366:565–569. - PubMed
-
- Boileau AJ, Kucken AM, Evers AR, Czajkowski C. Molecular dissection of benzodiazepine binding and allosteric coupling using chimeric gamma-aminobutyric acidA receptor subunits. Mol Pharmacol. 1998;53:295–303. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
