Base-pair substitutions in avian sarcoma virus U5 and U3 long terminal repeat sequences alter the process of DNA integration in vitro
- PMID: 11152486
- PMCID: PMC114019
- DOI: 10.1128/JVI.75.3.1132-1141.2001
Base-pair substitutions in avian sarcoma virus U5 and U3 long terminal repeat sequences alter the process of DNA integration in vitro
Abstract
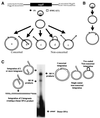
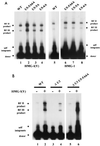
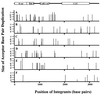
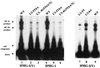
We have described a reconstituted avian sarcoma virus (ASV) concerted DNA integration system with specially designed mini-donor DNA containing a supF transcription unit, a supercoiled plasmid acceptor, purified bacterially expressed ASV integrase (IN), and human high-mobility-group protein I(Y). Integration in this system is dependent upon the mini-donor DNA having IN recognition sequences at both ends and upon both ends of the same donor integrating into the acceptor DNA. The integrated DNA product exhibits all of the features associated with integration of viral DNA in vivo (P. Hindmarsh et al., J. Virol., 73:2994-3003, 1999). Individual integrants are isolated from bacteria containing drug-resistant markers with amber mutations. This system was used to evaluate the importance of sequences in the terminal U5 and U3 long terminal repeats at positions 5 and/or 6, adjacent to the conserved CA dinucleotide. Base-pair substitutions introduced at these positions in U5 result in significant reductions in recovered integrants from bacteria, due to increases in one-ended insertion events. Among the recovered integrants from reactions with mutated U5 but not U3 IN recognition sequences were products that contain large deletions in the acceptor DNA. Base-pair substitutions at positions 5 and 6 in U3 mostly reduce the efficiency of integration of the modified donor. Together, these results indicate that sequences directly 5' to the conserved CA dinucleotide are very important for the process of concerted DNA integration. Furthermore, IN interacts with U3 and U5 termini differently, and aberrant end-processing events leading to nonconcerted DNA integration are more common in U5 than in U3.
Figures

Similar articles
-
Concerted integration of linear retroviral DNA by the avian sarcoma virus integrase in vitro: dependence on both long terminal repeat termini.J Virol. 1996 Jun;70(6):3571-80. doi: 10.1128/JVI.70.6.3571-3580.1996. J Virol. 1996. PMID: 8648691 Free PMC article.
-
Changes in the mechanism of DNA integration in vitro induced by base substitutions in the HIV-1 U5 and U3 terminal sequences.J Biol Chem. 2002 Mar 29;277(13):10938-48. doi: 10.1074/jbc.M108116200. Epub 2002 Jan 11. J Biol Chem. 2002. PMID: 11788585
-
Avian retrovirus U3 and U5 DNA inverted repeats. Role Of nonsymmetrical nucleotides in promoting full-site integration by purified virion and bacterial recombinant integrases.J Biol Chem. 1997 Sep 19;272(38):23938-45. doi: 10.1074/jbc.272.38.23938. J Biol Chem. 1997. PMID: 9295344
-
Retroviral DNA integration.Microbiol Mol Biol Rev. 1999 Dec;63(4):836-43, table of contents. doi: 10.1128/MMBR.63.4.836-843.1999. Microbiol Mol Biol Rev. 1999. PMID: 10585967 Free PMC article. Review.
-
Reconstitution of concerted DNA integration with purified components.Adv Virus Res. 1999;52:397-410. doi: 10.1016/s0065-3527(08)60308-5. Adv Virus Res. 1999. PMID: 10384244 Review. No abstract available.
Cited by
-
Biochemical and biophysical analyses of concerted (U5/U3) integration.Methods. 2009 Apr;47(4):229-36. doi: 10.1016/j.ymeth.2008.11.002. Epub 2008 Nov 29. Methods. 2009. PMID: 19049878 Free PMC article.
-
Subcellular localization and integration activities of rous sarcoma virus reverse transcriptase.J Virol. 2002 Jun;76(12):6205-12. doi: 10.1128/jvi.76.12.6205-6212.2002. J Virol. 2002. PMID: 12021354 Free PMC article.
-
Selection of functional mutations in the U5-IR stem and loop regions of the Rous sarcoma virus genome.BMC Biol. 2004 May 20;2:8. doi: 10.1186/1741-7007-2-8. BMC Biol. 2004. PMID: 15153244 Free PMC article.
-
Defining the DNA substrate binding sites on HIV-1 integrase.J Mol Biol. 2009 Jan 16;385(2):568-79. doi: 10.1016/j.jmb.2008.10.083. Epub 2008 Nov 7. J Mol Biol. 2009. PMID: 19014951 Free PMC article.
-
Recombinant human immunodeficiency virus type 1 integrase exhibits a capacity for full-site integration in vitro that is comparable to that of purified preintegration complexes from virus-infected cells.J Virol. 2005 Jul;79(13):8208-16. doi: 10.1128/JVI.79.13.8208-8216.2005. J Virol. 2005. PMID: 15956566 Free PMC article.
References
-
- Bustin M, Reeves R. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog Nucleic Acid Res Mol Biol. 1996;54:35–100. - PubMed
-
- Chow C S, Barnes C M, Lippard S J. A single HMG domain in high-mobility group 1 protein binds to DNAs as small as 20 base pairs containing the major cisplatin adduct. Biochemistry. 1995;34:2956–2964. - PubMed
-
- Farnet C M, Bushman F D. HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell. 1997;88:483–492. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

