Characterization of herpes simplex virus-containing organelles by subcellular fractionation: role for organelle acidification in assembly of infectious particles
- PMID: 11152497
- PMCID: PMC114030
- DOI: 10.1128/JVI.75.3.1236-1251.2001
Characterization of herpes simplex virus-containing organelles by subcellular fractionation: role for organelle acidification in assembly of infectious particles
Abstract
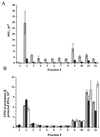
The cytoplasmic compartments occupied by exocytosing herpes simplex virus (HSV) are poorly defined. It is unclear which organelles contain the majority of trafficking virions and which are occupied by virions on a productive rather than defective assembly pathway. These problems are compounded by the fact that HSV-infected cells produce virus continuously over many hours. All stages in viral assembly and export therefore coexist, making it impossible to determine the sequence of events and their kinetics. To address these problems, we have established assays to monitor the presence of capsids and enveloped virions in cell extracts and prepared HSV-containing organelles from normally infected cells and from cells undergoing a single synchronized wave of viral egress. We find that, in both cases, HSV particles exit the nucleus and accumulate in organelles which cofractionate with the trans-Golgi network (TGN) and endosomes. In addition to carrying enveloped infectious virions in their lumen, HSV-bearing organelles also displayed nonenveloped capsids attached to their cytoplasmic surface. Neutralization of organellar pH by chloroquine or bafilomycin A resulted in the accumulation of noninfectious enveloped particles. We conclude that the organelles of the TGN/endocytic network play a key role in the assembly and trafficking of infectious HSV.
Figures





Similar articles
-
Progeny Varicella-Zoster Virus Capsids Exit the Nucleus but Never Undergo Secondary Envelopment during Autophagic Flux Inhibition by Bafilomycin A1.J Virol. 2019 Aug 13;93(17):e00505-19. doi: 10.1128/JVI.00505-19. Print 2019 Sep 1. J Virol. 2019. PMID: 31217243 Free PMC article.
-
Herpes Simplex Virus Capsid-Organelle Association in the Absence of the Large Tegument Protein UL36p.J Virol. 2015 Nov;89(22):11372-82. doi: 10.1128/JVI.01893-15. Epub 2015 Sep 2. J Virol. 2015. PMID: 26339048 Free PMC article.
-
Redistribution of cellular and herpes simplex virus proteins from the trans-golgi network to cell junctions without enveloped capsids.J Virol. 2004 Nov;78(21):11519-35. doi: 10.1128/JVI.78.21.11519-11535.2004. J Virol. 2004. PMID: 15479793 Free PMC article.
-
Virus Assembly and Egress of HSV.Adv Exp Med Biol. 2018;1045:23-44. doi: 10.1007/978-981-10-7230-7_2. Adv Exp Med Biol. 2018. PMID: 29896661 Review.
-
Morphology Remodeling and Selective Autophagy of Intracellular Organelles during Viral Infections.Int J Mol Sci. 2020 May 23;21(10):3689. doi: 10.3390/ijms21103689. Int J Mol Sci. 2020. PMID: 32456258 Free PMC article. Review.
Cited by
-
SARS-CoV-2, ACE2, and Hydroxychloroquine: Cardiovascular Complications, Therapeutics, and Clinical Readouts in the Current Settings.Pathogens. 2020 Jul 7;9(7):546. doi: 10.3390/pathogens9070546. Pathogens. 2020. PMID: 32645974 Free PMC article. Review.
-
Identification and characterization of the UL56 gene product of herpes simplex virus type 2.J Virol. 2002 Jul;76(13):6718-28. doi: 10.1128/jvi.76.13.6718-6728.2002. J Virol. 2002. PMID: 12050385 Free PMC article.
-
Structure and capsid association of the herpesvirus large tegument protein UL36.J Virol. 2010 Sep;84(18):9408-14. doi: 10.1128/JVI.00361-10. Epub 2010 Jul 14. J Virol. 2010. PMID: 20631146 Free PMC article.
-
Remodeling of host membranes during herpesvirus assembly and egress.Protein Cell. 2019 May;10(5):315-326. doi: 10.1007/s13238-018-0577-9. Epub 2018 Sep 21. Protein Cell. 2019. PMID: 30242641 Free PMC article. Review.
-
Deletion of the Pseudorabies Virus gE/gI-US9p complex disrupts kinesin KIF1A and KIF5C recruitment during egress, and alters the properties of microtubule-dependent transport in vitro.PLoS Pathog. 2020 Jun 8;16(6):e1008597. doi: 10.1371/journal.ppat.1008597. eCollection 2020 Jun. PLoS Pathog. 2020. PMID: 32511265 Free PMC article.
References
-
- Andoh T, Kawamata H, Umatake M, Terasawa K, Takegami T, Ochiai H. Effect of bafilomycin A1 on the growth of Japanese encephalitis virus in Vero cells. J Neurovirol. 1998;4:627–631. - PubMed
-
- Avitabile E, Ward P L, Di Lazzaro C, Torrisi M R, Roizman B, Campadelli-Fiume G. The herpes simplex virus UL20 protein compensates for the differential disruption of exocytosis of virions and viral membrane glycoproteins associated with fragmentation of the Golgi apparatus. J Virol. 1994;68:7397–7405. - PMC - PubMed
-
- Banfield W J, Kisch A L. The effect of chloroquine on herpesvirus infection in vitro and in vivo. Proc Soc Exp Biol Med. 1973;142:1018–1022. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

