Hepatitis C virus envelope protein E2 does not inhibit PKR by simple competition with autophosphorylation sites in the RNA-binding domain
- PMID: 11152499
- PMCID: PMC114032
- DOI: 10.1128/JVI.75.3.1265-1273.2001
Hepatitis C virus envelope protein E2 does not inhibit PKR by simple competition with autophosphorylation sites in the RNA-binding domain
Abstract
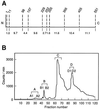
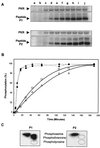
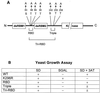
Double-stranded-RNA (dsRNA)-dependent protein kinase PKR is induced by interferon and activated upon autophosphorylation. We previously identified four autophosphorylated amino acids and elucidated their participation in PKR activation. Three of these sites are in the central region of the protein, and one is in the kinase domain. Here we describe the identification of four additional autophosphorylated amino acids in the spacer region that separates the two dsRNA-binding motifs in the RNA-binding domain. Eight amino acids, including these autophosphorylation sites, are duplicated in hepatitis C virus (HCV) envelope protein E2. This region of E2 is required for its inhibition of PKR although the mechanism of inhibition is not known. Replacement of all four of these residues in PKR with alanines did not dramatically affect kinase activity in vitro or in yeast Saccharomyces cerevisiae. However, when coupled with mutations of serine 242 and threonines 255 and 258 in the central region, these mutations increased PKR protein expression in mammalian cells, consistent with diminished kinase activity. A synthetic peptide corresponding to this region of PKR was phosphorylated in vitro by PKR, but phosphorylation was strongly inhibited after PKR was preincubated with HCV E2. Another synthetic peptide, corresponding to the central region of PKR and containing serine 242, was also phosphorylated by active PKR, but E2 did not inhibit this peptide as efficiently. Neither of the PKR peptides was able to disrupt the HCV E2-PKR interaction. Taken together, these results show that PKR is autophosphorylated on serine 83 and threonines 88, 89, and 90, that this autophosphorylation may enhance kinase activation, and that the inhibition of PKR by HCV E2 is not solely due to duplication of and competition with these autophosphorylation sites.
Figures




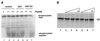
Similar articles
-
Uncoupling of RNA binding and PKR kinase activation by viral inhibitor RNAs.J Mol Biol. 2006 May 19;358(5):1270-85. doi: 10.1016/j.jmb.2006.03.003. Epub 2006 Mar 20. J Mol Biol. 2006. PMID: 16580685
-
Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein.Science. 1999 Jul 2;285(5424):107-10. doi: 10.1126/science.285.5424.107. Science. 1999. PMID: 10390359
-
HIV-I TAT inhibits PKR activity by both RNA-dependent and RNA-independent mechanisms.Arch Biochem Biophys. 2000 Jan 15;373(2):361-7. doi: 10.1006/abbi.1999.1583. Arch Biochem Biophys. 2000. PMID: 10620360
-
Activation of the RNA-dependent protein kinase (PKR) of lymphocytes by regulatory RNAs: implications for immunomodulation in HIV infection.Curr HIV Res. 2005 Oct;3(4):329-37. doi: 10.2174/157016205774370447. Curr HIV Res. 2005. PMID: 16250880 Review.
-
Inhibition of PKR by RNA and DNA viruses.Virus Res. 2006 Jul;119(1):100-10. doi: 10.1016/j.virusres.2005.10.014. Epub 2006 May 15. Virus Res. 2006. PMID: 16704884 Review.
Cited by
-
Analysis of monomeric and dimeric phosphorylated forms of protein kinase R.Biochemistry. 2010 Feb 16;49(6):1217-25. doi: 10.1021/bi901873p. Biochemistry. 2010. PMID: 20088595 Free PMC article.
-
Intrahepatic gene expression during chronic hepatitis C virus infection in chimpanzees.J Virol. 2004 Dec;78(24):13779-92. doi: 10.1128/JVI.78.24.13779-13792.2004. J Virol. 2004. PMID: 15564486 Free PMC article.
-
Viral determinants of resistance to treatment in patients with hepatitis C.Clin Microbiol Rev. 2007 Jan;20(1):23-38. doi: 10.1128/CMR.00010-06. Clin Microbiol Rev. 2007. PMID: 17223621 Free PMC article. Review.
-
Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins.J Virol. 2003 Jan;77(2):1501-11. doi: 10.1128/jvi.77.2.1501-1511.2003. J Virol. 2003. PMID: 12502864 Free PMC article.
-
Tinkering with translation: protein synthesis in virus-infected cells.Cold Spring Harb Perspect Biol. 2013 Jan 1;5(1):a012351. doi: 10.1101/cshperspect.a012351. Cold Spring Harb Perspect Biol. 2013. PMID: 23209131 Free PMC article. Review.
References
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. 2nd ed. Vol. 1. New York, N.Y: John Wiley & Sons; 1991.
-
- Baeuerle P A, Baltimore D. NF-kappa B: ten years after. Cell. 1996;87:13–20. - PubMed
-
- Bevilacqua P C, George C X, Samuel C E, Cech T R. Binding of the protein kinase PKR to RNAs with secondary structure defects: role of the tandem A-G mismatch and noncontiguous helixes. Biochemistry. 1998;37:6303–6316. - PubMed
-
- Brand S R, Kobayashi R, Mathews M B. The Tat protein of human immunodeficiency virus type 1 is a substrate and inhibitor of the interferon-induced, virally activated protein kinase, PKR. J Biol Chem. 1997;272:8388–8395. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

