Reovirus sigmaNS protein is required for nucleation of viral assembly complexes and formation of viral inclusions
- PMID: 11152519
- PMCID: PMC114052
- DOI: 10.1128/JVI.75.3.1459-1475.2001
Reovirus sigmaNS protein is required for nucleation of viral assembly complexes and formation of viral inclusions
Abstract
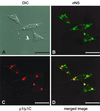
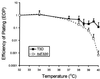
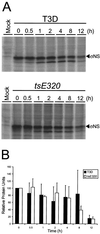
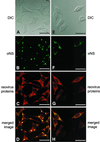
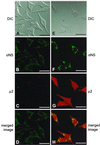
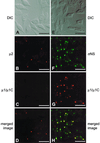
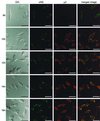
Progeny virions of mammalian reoviruses are assembled in the cytoplasm of infected cells at discrete sites termed viral inclusions. Studies of temperature-sensitive (ts) mutant viruses indicate that nonstructural protein sigmaNS and core protein mu2 are required for synthesis of double-stranded (ds) RNA, a process that occurs at sites of viral assembly. We used confocal immunofluorescence microscopy and ts mutant reoviruses to define the roles of sigmaNS and mu2 in viral inclusion formation. In cells infected with wild-type (wt) reovirus, sigmaNS and mu2 colocalize to large, perinuclear structures that correspond to viral inclusions. In cells infected at a nonpermissive temperature with sigmaNS-mutant virus tsE320, sigmaNS is distributed diffusely in the cytoplasm and mu2 is contained in small, punctate foci that do not resemble viral inclusions. In cells infected at a nonpermissive temperature with mu2-mutant virus tsH11.2, mu2 is distributed diffusely in the cytoplasm and the nucleus. However, sigmaNS localizes to discrete structures in the cytoplasm that contain other viral proteins and are morphologically indistinguishable from viral inclusions seen in cells infected with wt reovirus. Examination of cells infected with wt reovirus over a time course demonstrates that sigmaNS precedes mu2 in localization to viral inclusions. These findings suggest that viral RNA-protein complexes containing sigmaNS nucleate sites of viral replication to which other viral proteins, including mu2, are recruited to commence dsRNA synthesis.
Figures


References
-
- Ahmed R, Kauffman R S, Fields B N. Genetic variation during persistent reovirus infection: isolation of cold-sensitive and temperature-sensitive mutants from persistently infected L cells. Virology. 1983;131:71–78. - PubMed
-
- Antczak J B, Joklik W K. Reovirus genome segment assortment into progeny genomes studied by the use of monoclonal antibodies directed against reovirus proteins. Virology. 1992;187:760–776. - PubMed
-
- Coombs K M. Stoichiometry of reovirus structural proteins in virus, ISVP, and core particles. Virology. 1998;243:218–228. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

