The Max network gone mad
- PMID: 11154257
- PMCID: PMC86661
- DOI: 10.1128/MCB.21.3.691-702.2001
The Max network gone mad
Figures
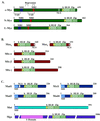
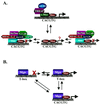


References
-
- Adams J M, Harris A W, Pinkert C A, Corcoran L M, Alexander W S, Palmiter R D, Brinster R L. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. - PubMed
-
- Alitalo K, Koskinen P, Makela T P, Saksela K, Sistonen L, Winqvist R. myc oncogenes: activation and amplification. Biochim Biophys Acta. 1987;907:1–32. - PubMed
-
- Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, DePinho R A. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. - PubMed
-
- Amati B, Brooks M W, Levy N, Littlewood T D, Evan G I, Land H. Oncogenic activity of the c-Myc protein requires dimerization with Max. Cell. 1993;72:233–245. - PubMed
-
- Amati B, Dalton S, Brooks M W, Littlewood T D, Evan G I, Land H. Transcriptional activation by the human c-Myc oncoprotein in yeast requires interaction with Max. Nature. 1992;359:423–426. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
