Molecular cloning and functional analysis of mouse C-terminal kinesin motor KifC3
- PMID: 11154264
- PMCID: PMC86668
- DOI: 10.1128/MCB.21.3.765-770.2001
Molecular cloning and functional analysis of mouse C-terminal kinesin motor KifC3
Abstract
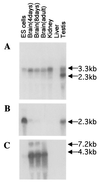
Proteins of the kinesin superfamily define a class of microtubule-dependent motors that play crucial roles in cell division and intracellular transport. To study the molecular mechanism of intracellular transport involving microtubule-dependent motors, a cDNA encoding a new kinesin-like protein called KifC3 was cloned from a mouse brain cDNA library. Sequence and secondary structure analysis revealed that KifC3 is a member of the C-terminal motor family. In contrast to other mouse C-terminal motors, KifC3 is apparently ubiquitous and may have a general role in intracellular transport. To understand the in vivo function of the KifC3 gene, we used homologous recombination in embryonic stem cells to construct knockout mouse strains for the KifC3 gene. Homozygous mutants of the KifC3 gene are viable, reproduce normally, and apparently develop normally. These results suggest that KifC3 is dispensable for normal development and reproduction in the mouse.
Figures



Similar articles
-
Functional analysis of mouse C-terminal kinesin motor KifC2.Mol Cell Biol. 2001 Apr;21(7):2463-6. doi: 10.1128/MCB.21.7.2463-2466.2001. Mol Cell Biol. 2001. PMID: 11259594 Free PMC article.
-
KIFC3, a microtubule minus end-directed motor for the apical transport of annexin XIIIb-associated Triton-insoluble membranes.J Cell Biol. 2001 Oct 1;155(1):77-88. doi: 10.1083/jcb.200108042. J Cell Biol. 2001. PMID: 11581287 Free PMC article.
-
Identification, partial characterization, and genetic mapping of kinesin-like protein genes in mouse.Genomics. 1997 Oct 1;45(1):123-31. doi: 10.1006/geno.1997.4901. Genomics. 1997. PMID: 9339368
-
Kinesin assembly and movement in cells.Annu Rev Biophys. 2011;40:267-88. doi: 10.1146/annurev-biophys-042910-155310. Annu Rev Biophys. 2011. PMID: 21332353 Review.
-
Structural links to kinesin directionality and movement.Nat Struct Biol. 2000 Jun;7(6):456-60. doi: 10.1038/75850. Nat Struct Biol. 2000. PMID: 10881190 Review.
Cited by
-
Emerging roles of centrosome cohesion.Open Biol. 2022 Oct;12(10):220229. doi: 10.1098/rsob.220229. Epub 2022 Oct 26. Open Biol. 2022. PMID: 36285440 Free PMC article. Review.
-
Identification of a novel gene fusion in ALT positive osteosarcoma.Oncotarget. 2018 Aug 28;9(67):32868-32880. doi: 10.18632/oncotarget.26029. eCollection 2018 Aug 28. Oncotarget. 2018. PMID: 30214690 Free PMC article.
-
Functional analysis of mouse C-terminal kinesin motor KifC2.Mol Cell Biol. 2001 Apr;21(7):2463-6. doi: 10.1128/MCB.21.7.2463-2466.2001. Mol Cell Biol. 2001. PMID: 11259594 Free PMC article.
-
Association of a nonmuscle myosin II with axoplasmic organelles.Mol Biol Cell. 2002 Mar;13(3):1046-57. doi: 10.1091/mbc.01-06-0315. Mol Biol Cell. 2002. PMID: 11907281 Free PMC article.
-
Role of KIFC3 motor protein in Golgi positioning and integration.J Cell Biol. 2002 Jul 22;158(2):293-303. doi: 10.1083/jcb.200202058. Epub 2002 Jul 22. J Cell Biol. 2002. PMID: 12135985 Free PMC article.
References
-
- Ando A, Kikuti Y Y, Kawata H, Okamoto N, Imai T, Eki T, Yokoyama K, Soeda E, Ikemura T, Abe K, et al. Cloning of a new kinesin-related gene located at the centromeric end of the human MHC region. Immunogenetics. 1994;39:194–200. - PubMed
-
- Bloom G S, Endow S A. Motor proteins 1: kinesins. Protein Profile. 1995;2:1105–1171. - PubMed
-
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. - PubMed
-
- Goldstein L S, Philp A V. The road less traveled: emerging principles of kinesin motor utilization. Annu Rev Cell Dev Biol. 1999;15:141–183. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
