Sequence-specific transcriptional repression by KS1, a multiple-zinc-finger-Krüppel-associated box protein
- PMID: 11154279
- PMCID: PMC86683
- DOI: 10.1128/MCB.21.3.928-939.2001
Sequence-specific transcriptional repression by KS1, a multiple-zinc-finger-Krüppel-associated box protein
Abstract
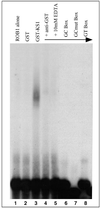
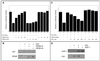
The vertebrate genome contains a large number of Krüppel-associated box-zinc finger genes that encode 10 or more C(2)-H(2) zinc finger motifs. Members of this gene family have been proposed to function as transcription factors by binding DNA through their zinc finger region and repressing gene expression via the KRAB domain. To date, however, no Krüppel-associated box-zinc finger protein (KRAB-ZFP) and few proteins with 10 or more zinc finger motifs have been shown to bind DNA in a sequence-specific manner. Our laboratory has recently identified KS1, a member of the KRAB-ZFP family that contains 10 different C(2)-H(2) zinc finger motifs, 9 clustered at the C terminus with an additional zinc finger separated by a short linker region. In this study, we used a random oligonucleotide binding assay to identify a 27-bp KS1 binding element (KBE). Reporter assays demonstrate that KS1 represses the expression of promoters containing this DNA sequence. Deletion and site-directed mutagenesis reveal that KS1 requires nine C-terminal zinc fingers and the KRAB domain for transcriptional repression through the KBE site, whereas the isolated zinc finger and linker region are dispensable for this function. Additional biochemical assays demonstrate that the KS1 KRAB domain interacts with the KAP-1 corepressor, and mutations that abolish this interaction alleviate KS1-mediated transcriptional repression. Thus, this study provides the first direct evidence that a KRAB-ZFP binds DNA to regulate gene expression and provides insight into the mechanisms used by multiple-zinc-finger proteins to recognize DNA sequences.
Figures






References
-
- Bellefroid E J, Lecocq P J, Benhida A, Poncelet D A, Belayew A, Martial J A. The human genome contains hundreds of genes coding for finger proteins of the Krüppel type. DNA. 1989;8:377–387. - PubMed
-
- Blackwell T K, Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990;250:1104–1110. - PubMed
-
- Cook T, Gebelein B, Mesa K, Mladek A, Urrutia R. Molecular cloning and characterization of TIEG2 reveals a new subfamily of transforming growth factor-beta-inducible Sp1-like zinc finger-encoding genes involved in the regulation of cell growth. J Biol Chem. 1998;273:25929–25936. - PubMed
-
- Cook T, Gebelein B, Urrutia R. Sp1 and its likes: biochemical and functional predictions for a growing family of zinc finger transcription factors. Ann N Y Acad Sci. 1999;880:94–102. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
