Identification of the maize amyloplast stromal 112-kD protein as a plastidic starch phosphorylase
- PMID: 11154342
- PMCID: PMC61015
- DOI: 10.1104/pp.125.1.351
Identification of the maize amyloplast stromal 112-kD protein as a plastidic starch phosphorylase
Abstract
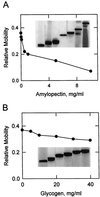
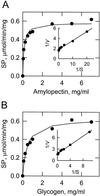
Amyloplast is the site of starch synthesis in the storage tissue of maize (Zea mays). The amyloplast stroma contains an enriched group of proteins when compared with the whole endosperm. Proteins with molecular masses of 76 and 85 kD have been identified as starch synthase I and starch branching enzyme IIb, respectively. A 112-kD protein was isolated from the stromal fraction by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and subjected to tryptic digestion and amino acid sequence analysis. Three peptide sequences showed high identity to plastidic forms of starch phosphorylase (SP) from sweet potato, potato, and spinach. SP activity was identified in the amyloplast stromal fraction and was enriched 4-fold when compared with the activity in the whole endosperm fraction. Native and sodium dodecyl sulfate-polyacrylamide gel electrophoresis analyses showed that SP activity was associated with the amyloplast stromal 112-kD protein. In addition, antibodies raised against the potato plastidic SP recognized the amyloplast stromal 112-kD protein. The amyloplast stromal 112-kD SP was expressed in whole endosperm isolated from maize harvested 9 to 24 d after pollination. Results of affinity electrophoresis and enzyme kinetic analyses showed that the amyloplast stromal 112-kD SP preferred amylopectin over glycogen as a substrate in the synthetic reaction. The maize shrunken-4 mutant had reduced SP activity due to a decrease of the amyloplast stromal 112-kD enzyme.
Figures



Similar articles
-
Purification and characterization of the maize amyloplast stromal 112-kDa starch phosphorylase.Arch Biochem Biophys. 2001 Apr 1;388(1):155-64. doi: 10.1006/abbi.2000.2267. Arch Biochem Biophys. 2001. PMID: 11361132
-
Polypeptides of the maize amyloplast stroma. Stromal localization of starch-biosynthetic enzymes and identification of an 81-kilodalton amyloplast stromal heat-shock cognate.Plant Physiol. 1998 Apr;116(4):1451-60. doi: 10.1104/pp.116.4.1451. Plant Physiol. 1998. PMID: 9536063 Free PMC article.
-
Isolation, identification and characterisation of starch-interacting proteins by 2-D affinity electrophoresis.Electrophoresis. 2006 May;27(9):1832-9. doi: 10.1002/elps.200500400. Electrophoresis. 2006. PMID: 16645949
-
Starch phosphorylase: role in starch metabolism and biotechnological applications.Crit Rev Biotechnol. 2009;29(3):214-24. doi: 10.1080/07388550902926063. Crit Rev Biotechnol. 2009. PMID: 19708823 Review.
-
Molecular Functions and Pathways of Plastidial Starch Phosphorylase (PHO1) in Starch Metabolism: Current and Future Perspectives.Int J Mol Sci. 2021 Sep 28;22(19):10450. doi: 10.3390/ijms221910450. Int J Mol Sci. 2021. PMID: 34638789 Free PMC article. Review.
Cited by
-
Understanding Starch Metabolism in Pea Seeds towards Tailoring Functionality for Value-Added Utilization.Int J Mol Sci. 2021 Aug 20;22(16):8972. doi: 10.3390/ijms22168972. Int J Mol Sci. 2021. PMID: 34445676 Free PMC article. Review.
-
CRISPR/Cas9-induced monoallelic mutations in the cytosolic AGPase large subunit gene APL2 induce the ectopic expression of APL2 and the corresponding small subunit gene APS2b in rice leaves.Transgenic Res. 2018 Oct;27(5):423-439. doi: 10.1007/s11248-018-0089-7. Epub 2018 Aug 11. Transgenic Res. 2018. PMID: 30099722
-
Responses to Hypoxia and Endoplasmic Reticulum Stress Discriminate the Development of Vitreous and Floury Endosperms of Conventional Maize (Zea mays) Inbred Lines.Front Plant Sci. 2017 Apr 13;8:557. doi: 10.3389/fpls.2017.00557. eCollection 2017. Front Plant Sci. 2017. PMID: 28450877 Free PMC article.
-
Gradually Decreasing Starch Branching Enzyme Expression Is Responsible for the Formation of Heterogeneous Starch Granules.Plant Physiol. 2018 Jan;176(1):582-595. doi: 10.1104/pp.17.01013. Epub 2017 Nov 13. Plant Physiol. 2018. PMID: 29133372 Free PMC article.
-
A translatome-transcriptome multi-omics gene regulatory network reveals the complicated functional landscape of maize.Genome Biol. 2023 Mar 29;24(1):60. doi: 10.1186/s13059-023-02890-4. Genome Biol. 2023. PMID: 36991439 Free PMC article.
References
-
- Akatsuka T, Nelson OE. Starch granule-bound adenosine diphosphate Glc-starch glucosyltransferases of maize seeds. J Biol Chem. 1966;241:2280–2286. - PubMed
-
- Ball S, Guan H, James M, Myers A, Keeling P, Mouille G, Buleon A, Colonna P, Preiss J. From glycogen to amylopectin: a model for the biogenesis of the plant starch granule. Cell. 1996;86:349–352. - PubMed
-
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein using the principles of protein dye-binding. Anal Biochem. 1976;72:248–254. - PubMed
-
- Burr B, Nelson OE. The phosphorylases of developing maize seeds. Ann NY Acad Sci. 1973;210:129–138. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

