Genomewide function conservation and phylogeny in the Herpesviridae
- PMID: 11156614
- PMCID: PMC311046
- DOI: 10.1101/gr.149801
Genomewide function conservation and phylogeny in the Herpesviridae
Abstract
The Herpesviridae are a large group of well-characterized double-stranded DNA viruses for which many complete genome sequences have been determined. We have extracted protein sequences from all predicted open reading frames of 19 herpesvirus genomes. Sequence comparison and protein sequence clustering methods have been used to construct herpesvirus protein homologous families. This resulted in 1692 proteins being clustered into 243 multiprotein families and 196 singleton proteins. Predicted functions were assigned to each homologous family based on genome annotation and published data and each family classified into seven broad functional groups. Phylogenetic profiles were constructed for each herpesvirus from the homologous protein families and used to determine conserved functions and genomewide phylogenetic trees. These trees agreed with molecular-sequence-derived trees and allowed greater insight into the phylogeny of ungulate and murine gammaherpesviruses.
Figures


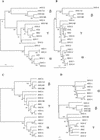
References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Andrade MA, Ouzounis C, Sander C, Tamames J, Valencia A. Functional classes in the three domains of life. J Mol Evol. 1999;49:551–557. - PubMed
-
- Benner SA, Cohen MA, Gonnet GH. Amino acid substitution during functionally constrained divergent evolution of protein sequences. Prot Eng. 1994;7:1323–1332. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
