Efficient male and female germline transmission of a human chromosomal vector in mice
- PMID: 11156621
- PMCID: PMC311020
- DOI: 10.1101/gr.159901
Efficient male and female germline transmission of a human chromosomal vector in mice
Abstract
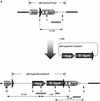
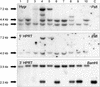
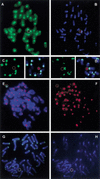
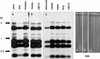
A small accessory chromosome that was mitotically stable in human fibroblasts was transferred into the hprt(-) hamster cell line CH and developed as a human chromosomal vector (HCV) by the introduction of a selectable marker and the 3' end of an HPRT minigene preceded by a loxP sequence. This HCV is stably maintained in the hamster cell line. It consists mainly of alphoid sequences of human chromosome 20 and a fragment of human chromosome region 1p22, containing the tissue factor gene F3. The vector has an active centromere, and telomere sequences are lacking. By transfecting a plasmid containing the 5' end of HPRT and a Cre-encoding plasmid into the HCV(+) hamster cell line, the HPRT minigene was reconstituted by Cre-mediated recombination and expressed by the cells. The HCV was then transferred to male mouse R1-ES cells and it did segregate properly. Chimeras were generated containing the HCV as an independent chromosome in a proportion of the cells. Part of the male and female offspring of the chimeras did contain the HCV. The HCV(+) F1 animals harbored the extra chromosome in >80% of the cells. The HCV was present as an independent chromosome with an active centromere and the human F3 gene was expressed from the HCV in a human-tissue-specific manner. Both male and female F1 mice did transmit the HCV to F2 offspring as an independent chromosome with properties similar to the original vector. This modified small accessory chromosome, thus, shows the properties of a useful chromosomal vector: It segregates stably as an independent chromosome, sequences can be inserted in a controlled way and are expressed from the vector, and the HCV is transmitted through the male and female germline in mice.
Figures


Similar articles
-
Tissue-specific expression of a BAC transgene targeted to the Hprt locus in mouse embryonic stem cells.Genomics. 2004 Jun;83(6):1072-82. doi: 10.1016/j.ygeno.2003.12.015. Genomics. 2004. PMID: 15177560
-
Mouse embryonic stem cells with a multi-integrase mouse artificial chromosome for transchromosomic mouse generation.Transgenic Res. 2015 Aug;24(4):717-27. doi: 10.1007/s11248-015-9884-6. Epub 2015 Jun 9. Transgenic Res. 2015. PMID: 26055730 Free PMC article.
-
Engineering chromosomal rearrangements in mice.Nat Rev Genet. 2001 Oct;2(10):780-90. doi: 10.1038/35093564. Nat Rev Genet. 2001. PMID: 11584294 Review.
-
HPRT minigene generates chimeric transcripts as a by-product of gene targeting.Genesis. 2007 May;45(5):275-81. doi: 10.1002/dvg.20300. Genesis. 2007. PMID: 17457929
-
Gene targeting in murine embryonic stem cells: introduction of specific alterations into the mammalian genome.Genet Anal Tech Appl. 1990 Dec;7(8):219-27. doi: 10.1016/0735-0651(90)90004-y. Genet Anal Tech Appl. 1990. PMID: 2091698 Review.
Cited by
-
Increased missegregation and chromosome loss with decreasing chromosome size in vertebrate cells.Chromosoma. 2006 Feb;115(1):60-74. doi: 10.1007/s00412-005-0032-6. Epub 2005 Nov 3. Chromosoma. 2006. PMID: 16267674
-
Novel method to load multiple genes onto a mammalian artificial chromosome.PLoS One. 2014 Jan 15;9(1):e85565. doi: 10.1371/journal.pone.0085565. eCollection 2014. PLoS One. 2014. PMID: 24454889 Free PMC article.
-
A novel system for simultaneous or sequential integration of multiple gene-loading vectors into a defined site of a human artificial chromosome.PLoS One. 2014 Oct 10;9(10):e110404. doi: 10.1371/journal.pone.0110404. eCollection 2014. PLoS One. 2014. PMID: 25303219 Free PMC article.
-
A mammalian artificial chromosome engineering system (ACE System) applicable to biopharmaceutical protein production, transgenesis and gene-based cell therapy.Nucleic Acids Res. 2004 Dec 7;32(21):e172. doi: 10.1093/nar/gnh169. Nucleic Acids Res. 2004. PMID: 15585659 Free PMC article.
-
Telomere-independent homologue pairing and checkpoint escape of accessory ring chromosomes in male mouse meiosis.J Cell Biol. 2003 Sep 1;162(5):795-807. doi: 10.1083/jcb.200305065. J Cell Biol. 2003. PMID: 12952934 Free PMC article.
References
-
- Au HC, Mascarello JT, Scheffler IE. Targeted integration of a dominant neo(R) marker into a 2- to 3-Mb human minichromosome and transfer between cells. Cytogenet Cell Genet. 1999;86:194–203. - PubMed
-
- Baldini A, Archidiacono N, Carbone R, Bolino A, Shridhar V, Miller OJ, Miller DA, Ward DC, Rocchi M. Isolation and comparative mapping of a human chromosome 20-specific alpha-satellite DNA clone. Cytogenet Cell Genet. 1992;59:12–16. - PubMed
-
- Brown KE, Barnett MA, Burgtorf C, Shaw P, Buckle VJ, Brown WR. Dissecting the centromere of the human Y chromosome with cloned telomeric DNA. Hum Mol Genet. 1994;3:1227–1237. - PubMed
-
- Burgoyne PS, Mahadevaiah SK. Unpaired sex chromosomes and gametogenetic failure. In: Summers AT, Chandley AC, editors. Chromosomes Today, Volume 11. New York: Chapman and Hall; 1993. pp. 243–263.
-
- Choi TK, Hollenbach PW, Pearson BE, Ueda RM, Weddell GN, Kurahara CG, Woodhouse CS, Kay RM, Loring JF. Transgenic mice containing a human heavy chain immunoglobulin gene fragment cloned in a yeast artificial chromosome. Nat Genet. 1993;4:117–123. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
