Binding of C4b-binding protein to porin: a molecular mechanism of serum resistance of Neisseria gonorrhoeae
- PMID: 11157049
- PMCID: PMC2195916
- DOI: 10.1084/jem.193.3.281
Binding of C4b-binding protein to porin: a molecular mechanism of serum resistance of Neisseria gonorrhoeae
Abstract
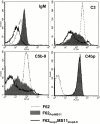
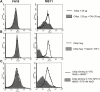
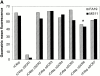
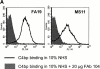
We screened 29 strains of Neisseria gonorrhoeae and found 16/21 strains that resisted killing by normal human serum and 0/8 serum sensitive strains that bound the complement regulator, C4b-binding protein (C4bp). Microbial surface-bound C4bp demonstrated cofactor activity. We constructed gonococcal strains with hybrid porin (Por) molecules derived from each of the major serogroups (Por1A and Por1B) of N. gonorrhoeae, and showed that the loop 1 of Por1A is required for C4bp binding. Por1B loops 5 and 7 of serum-resistant gonococci together formed a negatively charged C4bp-binding domain. C4bp-Por1B interactions were ionic in nature (inhibited by high salt or by heparin), whereas the C4bp-Por1A bond was hydrophobic. Only recombinant C4bp mutant molecules containing the NH2-terminal alpha-chain short consensus repeat (SCR1) bound to both Por1A and Por1B gonococci, suggesting that SCR1 contained Por binding sites. C4bp alpha-chain monomers did not bind gonococci, indicating that the polymeric form of C4bp was required for binding. Using fAb fragments against C4bp SCR1, C4bp binding to Por1A and Por1B strains was inhibited in a complement-dependent serum bactericidal assay. This resulted in complete killing of these otherwise fully serum resistant strains in only 10% normal serum, underscoring the importance of C4bp in mediating gonococcal serum resistance.
Figures
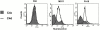
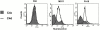







References
-
- Rice P.A., McCormack W.M., Kasper D.L. Natural serum bactericidal activity against Neisseria gonorrhoeae isolates from disseminated, locally invasive, and uncomplicated disease. J. Immunol. 1980;124:2105–2109. - PubMed
-
- Schumacher G.F. Immunology of spermatozoa and cervical mucus. Hum. Reprod. 1988;3:289–300. - PubMed
-
- Price R.J., Boettcher B. The presence of complement in human cervical mucus and its possible relevance to infertility in women with complement-dependent sperm-immobilizing antibodies. Fertil. Steril. 1979;32:61–66. - PubMed
-
- Bischof P., Planas-Basset D., Meisser A., Campana A. Investigations on the cell type responsible for the endometrial secretion of complement component 3 (C3) Hum. Reprod. 1994;9:1652–1659. - PubMed
-
- Sayegh R.A., Tao X.J., Awwad J.T., Isaacson K.B. Localization of the expression of complement component 3 in the human endometrium by in situ hybridization. J. Clin. Endocrinol. Metab. 1996;81:1641–1649. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

